In the current issue of Blood, Yokoyama et al1 demonstrate that an IL7R mutation similar to those found in patients with acute lymphoblastic leukemia (ALL)2-4 can be leukemogenic in vivo when expressed in normal hematopoietic progenitors.
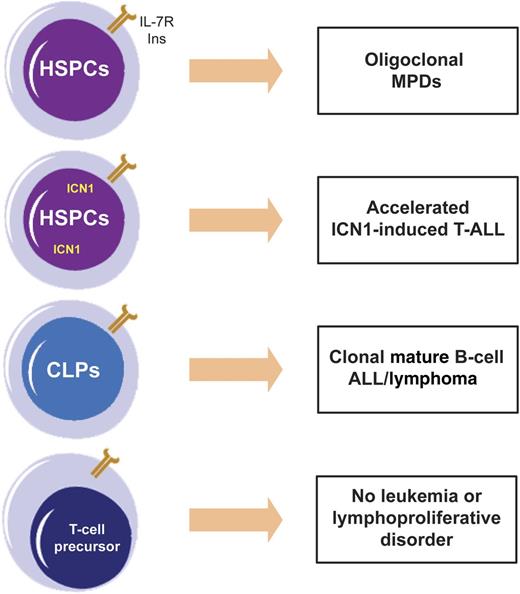
Effects of IL-7Rα gain-of-function mutation (INS) are context dependent. Ectopic expression of mutant IL-7Rα alone in HSPCs originates oligoclonal myeloproliferative diseases (MPDs); when combined with constitutively active ICN1, it accelerates T-ALL; when expressed in CLPs, it drives mature B-cell ALL/lymphoma. Surprisingly, expression of INS in T-cell progenitors did not induce T-cell leukemia or lymphoproliferation.
Effects of IL-7Rα gain-of-function mutation (INS) are context dependent. Ectopic expression of mutant IL-7Rα alone in HSPCs originates oligoclonal myeloproliferative diseases (MPDs); when combined with constitutively active ICN1, it accelerates T-ALL; when expressed in CLPs, it drives mature B-cell ALL/lymphoma. Surprisingly, expression of INS in T-cell progenitors did not induce T-cell leukemia or lymphoproliferation.
Interleukin 7 (IL-7), a cytokine produced in the bone marrow, thymus, and other organs, is absolutely required for T-cell development and homeostasis.5 Similar to the ligand, genetic inactivation of any of the 2 subunits of the IL-7 receptor (IL-7R), a heterodimer of IL-7Rα (encoded by the IL7R gene) and γc, leads to severe immunodeficiency. The notable capacities of IL-7 and IL-7R to stimulate T cells have driven phase I clinical trials to evaluate the value of recombinant IL-7 as a booster of immune reconstitution in the context of cancer therapy.5 However, what if these 2 partners had a Mr Hyde facet hiding behind their kind Dr Jekyll nature? Well, it appears they do. Like a mirror image of immunodeficiency due to their inactivation, all evidence indicates that excessive signaling via IL-7 and IL-7R can contribute to leukemia progression.6 Compellingly, 9% to 10% of T-cell ALL (T-ALL) and <1% of B-cell ALL patients display IL7R gain-of-function mutations, which drive constitutive activation of downstream signaling and cell transformation in vitro.2-4 What about in vivo? IL-7Rα mutants, but not the wild-type receptor, were shown to induce tumorigenesis when expressed in D1 p53-null mouse thymocytes subcutaneously transplanted into recipient mice.2 However useful, this is an artificial model that does not evaluate the impact of IL7R gain-of-function genetic lesions in the scenario where they actually occur: during hematopoiesis.
In their study, Yokoyama and colleagues demonstrate for the first time that 1 of these mutations, identified in the T-ALL cell line DND-41,7 can be leukemogenic in vivo when retrovirally transduced hematopoietic stem and progenitor cells (HSPCs) are transplanted into recipient mice. Perhaps their most remarkable observation is that the transforming ability of mutant IL-7Rα, and the phenotype elicited by it, depended on the developmental stage/lineage in which the mutation was expressed (see figure). Ectopic expression of mutant IL-7Rα in HSPCs caused oligoclonal myeloproliferative diseases (MPDs), whereas forced expression in common lymphoid progenitors (CLPs) originated mature B-cell acute leukemia/lymphoma. By contrast, transduction of CD4 CD8 double-negative T-cell progenitors did not induce T-cell lymphoproliferative disease or leukemia. However, when coexpressed in HSPCs, the mutant receptor significantly accelerated the aggressive T-ALL that is driven by constitutively active intracellular Notch1 (ICN1).
There are several intriguing features in these twists and turns. It is evident that myeloid lineage precursors displayed a considerable selective advantage as a consequence of ectopic IL-7Rα constitutive signals. However, the same signals that originated myeloproliferative disorders did not appear to be sufficient to initiate full-blown leukemia. In humans, one may speculate that the probability of IL7R mutations occurring in myeloid neoplasms should be low, especially considering that 2 hits affecting the IL7R locus must occur: one leading to aberrant expression and a second inducing mutational activation of IL-7Rα. Nonetheless, whether human myeloid neoplasms of any kind display driver IL7R mutations remains to be determined. Also unforeseen was the fact that mature B-cell ALL/lymphoma instead of B-cell precursor ALL, where the mutations have been identified in humans, arose as a consequence of mutant IL-7Rα expression in CLPs. The real surprise, however, came from the realization that expression of mutant IL-7Rα alone in T-cell progenitors did not drive T-cell leukemia >3 months after transplantation into recipient mice. Knowing that IL7R mutations are far more frequent in T-cell than B-cell ALL, this is unexpected. The authors argue it may be due in part to limited engraftment of transduced T-cell progenitors. Or perhaps, one could further speculate that T-cell tumors promoted by IL-7R activation arise with long latency, similar to TAL1-driven leukemia in mice, for instance. If this is true, it could be suggestive of the necessity for subsequent oncogenic hits to complement the transforming effects of IL7R mutations. In this regard, it will be relevant to explore possible synergisms between IL-7Rα activation and weak NOTCH1 mutant alleles commonly found in T-ALL patients, which contrast with ICN1 overexpression in that they do not initiate leukemia per se but cooperate with other oncogenes to accelerate leukemia onset.8 Likewise, evaluating the in vivo cooperation between mutation of IL-7Rα and HOXA gene deregulation would merit investigation, given that IL7R mutations are enriched in T-ALL cases with HOXA aberrations.2 Finally, given the presence of IL7R mutations in early T-cell precursor ALL (ETP-ALL),4 a very poor prognosis T-ALL subgroup, it would be of considerable value if genetic interactions between IL-7R mutational activation and other oncogenes were identified that drive ETP-ALL in vivo.
What are the specific mediators of mutant IL7R oncogenic effects in the different hematopoietic populations? To try and answer this question, the authors performed comparative gene expression profiling. They found several genes whose differential expression between the wild-type and mutant receptor is consistent with potential roles as effectors of IL-7R–dependent transformation, namely HES1 in MPD, PIM1 in B-cell ALL/lymphoma, and IGFR1 in ICN1-triggered T-ALL. Whether these genes are actually involved in IL7R pathophysiology in the different scenarios will require future validation. Interestingly, gene set enrichment analysis further suggested that there may be an interferon gene signature underlying IL7R oncogenic effects. This may have clinical implications, because IL7R mutant ALL cells could, in theory, be susceptible to the antiproliferative effects of type I interferon, similar to mutant JAK1 cases that present a comparable signature.9
The complexities of IL-7R involvement in leukemogenesis are only beginning to be exposed. For instance, there is evidence that IL-7Rα deficiency in p53-null mice potentiates lymphomagenesis by exacerbating telomere erosion in T-cell precursors.10 Would such a mechanism apply to humans? There are no reported loss-of-function mutations or deletions of the IL7R locus in human ALL patients, but as the evidence for the involvement of IL-7 and its receptor in cancer progression steadily extends beyond leukemia,6 it will be exciting to follow the twists and turns of Dr Jekyll and Mr Hyde and see where they will lead.
Conflict-of-interest disclosure: The author declares no competing financial interests.