In this issue of Blood, Choi et al report a 3-dimensional (3D) reconstruction model of integrin αIIbβ3 in its latent state which challenges the existing paradigm and provides new insights into the mechanism of integrin activation.1
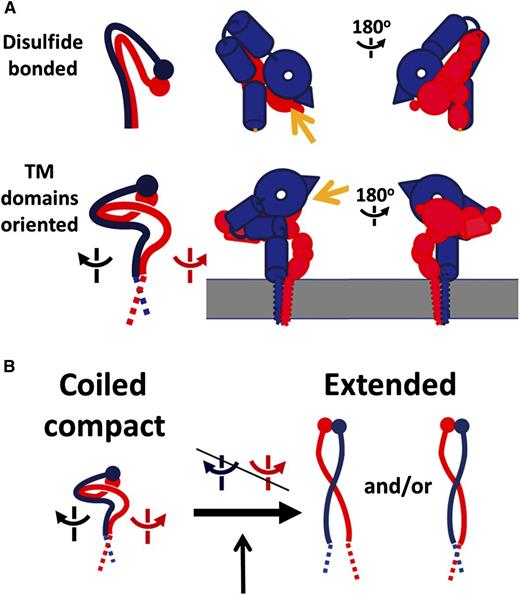
Changed interface between TM domains by cytosolic adaptors during inside-out signaling. The heterodimeric (α/β) integrins belong to a family of 24 members composed of different α/β subunit combinations. Each subunit consists of a large extracellular ligand-binding domain (ectodomain), a single transmembrane domain, and a small cytoplasmic tail of ∼20 to 70 residues. (A) Comparison of 2 different models of inactive integrin. (Top panel) Overall topology of integrin αIIbβ3 ectodomain based on crystallographic information. The ligand-binding head piece points down toward the membrane. αIIb is colored in blue and β3 in red. (Bottom panel) Overall topology of intact integrin αIIbβ3 based on the EM study.1 Yellow arrows indicate the ligand-binding site. (B) A modified switchblade conformational transition model for integrin activation. See Figure 4 in the article by Choi et al that begins on page 4165.
Changed interface between TM domains by cytosolic adaptors during inside-out signaling. The heterodimeric (α/β) integrins belong to a family of 24 members composed of different α/β subunit combinations. Each subunit consists of a large extracellular ligand-binding domain (ectodomain), a single transmembrane domain, and a small cytoplasmic tail of ∼20 to 70 residues. (A) Comparison of 2 different models of inactive integrin. (Top panel) Overall topology of integrin αIIbβ3 ectodomain based on crystallographic information. The ligand-binding head piece points down toward the membrane. αIIb is colored in blue and β3 in red. (Bottom panel) Overall topology of intact integrin αIIbβ3 based on the EM study.1 Yellow arrows indicate the ligand-binding site. (B) A modified switchblade conformational transition model for integrin activation. See Figure 4 in the article by Choi et al that begins on page 4165.
Integrins are a group of heterodimeric (α/β) transmembrane receptors crucial for mediating a variety of cell adhesion–dependent physiological and pathological processes such as tissue formation, blood clotting, immune responses, and tumor metastasis.2 Discovered 3 decades ago, integrins have been extensively studied with >56 000 citations found in PubMed. A central topic of the integrin research has been focused on understanding how integrin/ligand interaction (integrin affinity) is regulated. The answer to this question is crucial not only for understanding diverse integrin-mediated biological processes but also for diagnosing/treating various integrin-related diseases. A powerful way to address this question is to obtain detailed 3D structures of inactive integrin vs active integrin. In early 2000, the first groundbreaking crystal structure of integrin αvβ3 ectodomain was reported,3 which revealed a surprising bent conformation in which the ligand-binding head domain pointed down toward the membrane surface and the 2 legs were straight, parallel, and adjacent (see figure, panel A). A subsequent low-resolution cryo–electron microscopy (EM) model of intact integrin αIIbβ34 in detergent indicated a different ectodomain orientation, especially with the head pointing away from membrane, suggesting that the transmembrane-cytoplasmic domain may influence the global architecture of the receptor. Nevertheless, the same unusual bent fold was observed in several other integrin ectodomains including αIIbβ3 (for review, see Campbell and Humphries5 ). These ectodomain structures, together with a series of biochemical, biophysical, and cell biology studies, led to a widely accepted model that integrins are activated via a switchblade-like conformational transition.5 Meanwhile, structures of integrin αIIbβ3 transmembrane-cytoplasmic or cytoplasmic heterodimer were also pursued to understand how these segments control the resting, inactive state of the receptor.5 Nuclear Magnetic Resonance (NMR) analysis indicated that integrins contain a conserved α/β transmembrane interface but highly flexible cytoplasmic tail conformation. Notably, the membrane-proximal αIIb region was found to adopt helical,6 reverse turn,7 or disordered conformation.8 The C-terminal β3 cytoplasmic tail was also found to adopt variable conformations including helix, β-strand, or loop.6,8,9 Such structural variations may be caused by multiple factors such as different membrane-mimetic conditions, truncation of protein constructs, or binding to different partners/regulators. They may also dictate different functional or intermediate states of the receptor, which remains to be further investigated.
Given the uncertainties in structure and orientation of isolated integrin domains, Choi et al decided to pursue the structural investigation of full-length integrin. They purified intact inactive integrin αIIbβ3: the major platelet integrin that has been extensively studied for understanding the integrin structure/function. Because determining the structure of full-length integrin is still technically limited for x-ray crystallography and NMR, the authors used transmission EM to analyze the entire αIIbβ3 that was embedded in membrane-like nanodisc. By fitting the crystal structures of individual subdomains into an EM map, they were able to obtain the 3D reconstruction model of the ectodomain at 20.5Å resolution. The results were surprising: they found that the head domain points away from the membrane bilayer surface, contrasting to what has been posited from the crystal structure of the ectodomain (see figure). The upward orientation of the αIIbβ3 head domain was further confirmed by the antibody-based epitope mapping and is also consistent with the previous cryo-EM model of intact integrin αIIbβ3.4 The lower legs connecting to the transmembrane domain were coiled, which were also significantly different from the straight conformation in the crystal structure. This coiled conformation is likely the underlying basis to allow the upward orientation of the head domain. Based on these data, Choi et al proposed a modified switchblade model for integrin activation: rather than going through a bent-to-extended change in a single fulcrum, integrin ectodomain undergoes multiple interdomain movements as initiated by the transmembrane-cytoplasmic domain, which ultimately leads to an extended/active conformation of the receptor (see figure).
The resolution of the new αIIbβ3 model was not sufficient to resolve the transmembrane-cytoplasmic domain and hence continued effort is needed to overcome the technical limit. However, the results derived from this study are already of significant value to the integrin field as well as to membrane protein structural biology in general. First, the new αIIbβ3 model challenges the existing paradigm for the inactive integrin ectodomain conformation and orientation relative to membrane, leading to a revised model for the integrin activation. Second, the study emphasizes the importance of the transmembrane domain in influencing the global structure and orientation of any transmembrane protein. Because many ectodomain structures of transmembrane receptors have been deposited in the Protein Data Bank and more are being determined, the Choi et al study calls upon cautious interpretation of these structures. In the case of αIIbβ3, it is clear that the transmembrane domain was crucial for influencing the lower leg conformation and ultimately the upward orientation of the head domain. On the other hand, it should be pointed out that the Choi et al study was performed in the absence of extracellular ligands and intracellular regulators. It remains to be further investigated, hopefully at higher resolution, if integrin does undergo the proposed conformational transition upon binding to physiological ligands such as fibrinogen or intracellular activators such as talin and kindlin. It also remains to be seen whether integrin maintains the proposed inactive conformation in the presence of intracellular inhibitors such as calcium integrin-binding protein, Sharpin, and filamin.10 With the rapid improvement of biotechnology, such as protein sample preparation and high-resolution structural biology equipment, one may expect additional breakthroughs in the near future, which will further advance our understanding of the integrin function in diverse biological processes and hopefully also promote the development of better therapy for treating integrin-dependent human diseases.
Conflict-of-interest disclosure: The authors declare no competing financial interests.