Key Points
IL-15 has been implicated in CNS disease and leukemogenesis, but the biological mechanisms are unknown.
IL-15 increases pre-B ALL growth and upregulates CNS homing molecules, and MEK/ERK, PI3K, and NF-κB inhibitors block IL-15 growth effects.
Abstract
Genome-wide association studies have consistently implicated the interleukin-15 (IL-15) gene in acute lymphoblastic leukemia (ALL) biology, including associations with disease susceptibility, and increased risk of central nervous system (CNS) involvement. However, whether pre-B ALL blasts directly respond to IL-15 is unknown. Here, we show that most pre-B ALL primary samples and cell lines express IL-15 and components of its receptor and that primary pre-B ALL cells show increased growth in culture in response to IL-15. Investigation of mechanisms of action using IL-15–responsive SD-1 cells shows this growth advantage is maximal under low-serum conditions, mimicking those found in cerebrospinal fluid. IL-15 also upregulates PSGL-1 and CXCR3, molecules associated with CNS trafficking. Investigation of downstream signaling pathways indicates that IL-15 induces signal transducer and activator of transcription 5 (STAT5), extracellular signal-regulated kinase (ERK) 1/2, and to a lesser extent phosphatidylinositol 3-kinase (PI3K) and nuclear factor κB (NF-κB) phosphorylation. The IL-15–mediated growth advantage is abolished by mitogen-activated protein kinase kinase/extracellular signal-regulated kinase (MEK/ERK), PI3K, and NF-κB inhibitors but preserved in the presence of STAT5 inhibition. Together, these observations provide a mechanistic link between increased levels of IL-15 expression and leukemogenesis, high-risk disease, and CNS relapse and suggest potential therapeutic targets.
Introduction
Large-scale unbiased genomic approaches are increasingly used in leukemia research to identify factors that influence tumorigenesis, biological features, and/or response to therapy. Genetic “hits” identified by these investigations may be directly involved in the disease or bystanders incidentally linked to other (often unknown) genetic loci that drive the process. For this reason, it is important to verify the biological role of implicated genes if progress is to be made. Several large observational studies have linked interleukin-15 (IL-15) single-nucleotide polymorphisms (SNPs) or mRNA levels to aspects of acute lymphoblastic leukemia (ALL) biology. Two independent genome-wide association studies (GWAS) identified polymorphisms in the IL-15 gene as predictors of leukemia development1 and resistance to initial therapy.2 Microarray data suggest that IL-15 mRNA levels predict the likelihood of central nervous system (CNS) relapse in childhood ALL.3 Finally, a clinical study looking at IL-15 mRNA in adult ALL showed that higher IL-15 expression was associated with mediastinal and lymph node (LN) infiltration but not hepatosplenomegaly (rates of CNS involvement were not reported).4 Importantly, some IL-15 SNPs correlate with increased promoter activity when tested in reporter constructs,5 suggesting a possible direct link to pathogenesis. However, the mechanisms underlying any biological advantage for high IL-15 expression in ALL are currently unknown.
IL-15 is a pleiotropic cytokine sharing structural homology and receptor components with IL-26,7 ; together, these cytokines play pivotal roles in regulatory and effector functions of the normal immune system. IL-15 influences proliferation, differentiation, resistance to apoptosis,8 and cellular localization of T9 and B lymphocytes,10 neutrophils,11 and natural killer (NK) cells.12 A possible role for IL-15 in malignant disorders of the immune system was first suggested by its identification in an adult T-cell leukemia-lymphoma cell line13 and the fact that IL-15 transgenic mice develop an NK/T large granular lymphocyte leukemia-like disease.14,15 IL-15 stimulates proliferation of primary T-ALL blasts in culture,16 and chronic exposure of normal large granular lymphocyte leukemias to IL-15 induces leukemic transformation.17 Links to B-cell malignancies (myeloma18 and chronic lymphocytic leukemia19,20 ) have also been reported.
The IL-15 gene is expressed in many tissues, including the CNS.21,22 Two IL-15 isoforms encode the same mature IL-15 protein but with different signal peptides determining cellular localization.23 The 21-amino-acid short-signal peptide isoform (SSP-IL-15) is targeted to the cytoplasm; its function remains largely unknown. In contrast, the 48-amino-acid long-signal peptide isoform (LSP-IL-15) targets IL-15 for presentation on the cell surface and/or secretion. IL-15 signals via a heterotrimeric receptor complex comprising a common γ chain, an IL-2/IL-15Rβ chain,6 and a specific IL-15Rα chain.24 This IL-15Rα chain binds IL-15 with high affinity (1000-fold higher than IL-2/IL-2Rα interactions24 ) and can be secreted or membrane bound. IL-15/IL-15Rα heterodimers may be the main active form of IL-15 in human serum.25 Thus, IL-15 can be presented in cis or trans and act in an autocrine, paracrine, or juxtacrine fashion. It is unknown whether the reported associations between IL-15 SNPs and ALL reflect a direct biological role for IL-15 in ALL. If so, this could be due to direct effects of IL-15 on leukemic blasts or indirect effects via IL-15 modulation of the immune system or a combination of the 2.
In this study, we investigate the biological effect of IL-15 in pre-B ALL. We show that IL-15 is expressed by pre-B ALL blasts, which respond to both autocrine and paracrine IL-15 signaling. We show that the growth advantage of leukemic blasts exposed to IL-15 is maximal under low-serum conditions and identify downstream pathways responsible for the growth advantage. Thus, we have identified possible therapeutic targets that could be used to treat high-risk disease and/or CNS relapse.
Materials and methods
Cell culture and primary cells
Human ALL cell lines SD-1, REH, Molt-4, Sup-B15, SEM, and CCRF-CEM were maintained in RPMI supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen). Pre-B ALL primary samples were obtained from the Leukaemia Lymphoma Research Childhood Leukaemia Cellbank and from our local institutions. Cells used for receptor expression analysis were cultured in complete RPMI, at 37°C, 5% CO2 for 24 hours before harvesting in TRIzol (Invitrogen). Primary cells were processed and cryopreserved as previously described.26 For growth curves, primary cells were thawed and set up in suspension culture in HP01 media (Macopharma) supplemented with rhIL-3 (20 ng/mL), rhIL-7 (20 ng/mL), and rhSCF (50 ng/mL) (3-cytokine mix) (all R&D Systems).27 After 7 to 14 days in culture (recovery time), these cells were washed and split in to 2 conditions: 3-cytokine mix only or 3-cytokine mix + rhIL-15 (25 ng/mL) (Peprotech). Cells were maintained over a 3-week period with weekly half-media changes. Viability and absolute cell counts (live lymphoid blasts) were determined using a MACSQuant flow cytometer (Miltenyi Biotec) after staining with 7-aminoactinomycin-D (Invitrogen). Patient details are given in supplemental Table 1, available on the Blood Web site. This project was approved by the West of Scotland Research Ethics Committee. This study was conducted in accordance with the Declaration of Helsinki.
Xenotransplants
Nonobese diabetic Cg-PrkdcscidIl2rgtm1Wjl/SzJ breeders were obtained from Charles River, Europe and the colony maintained at the Central Research Facility, University of Glasgow. Mice were kept in sterile isolators with autoclaved food, bedding, and water. At 6 to 8 weeks of age, mice were injected intravenously with 1 × 106 leukemic cells via tail vein. Mice were euthanized at 6 weeks postinjection or earlier if unwell. All animal experiments were approved by the University of Glasgow Ethical Review Process Committee.
Histology
Murine brains were fixed in 10% neutral-buffered formalin and paraffin embedded. Hematoxylin and eosin staining (Sigma-Aldrich) was performed on 5-µm brain sections. Anti-CD45 immunohistochemistry used standard protocols; paraffin-embedded sections were dewaxed and hydrated prior to antigen retrieval at 100°C for 15 minutes in 0.01 mol/L citrate buffer (pH 6.0). Following blocking in 20% horse serum/phosphate-buffered saline–Tween 20 (0.05%) with avidin block, sections were incubated with mouse anti-human CD45 antibody/isotype control antibody (mouse IgG1) (Dako) and horse, biotinylated anti-mouse secondary antibody (Vector Laboratories) with biotin block. Sections were treated with ABC kit (Vector Laboratories) and DAB to visualize antibody binding. All imaging was performed on an Axiostar plus microscope, and images were acquired using Axiovision Rel 4.2 software (Carl Zeiss, Oberkochen, Germany).
PCR
Total RNA was extracted using an RNeasy kit (Qiagen) or following homogenization in TRIzol. All samples underwent DNase digestion (RNase-free DNase, Qiagen). RNA (200 ng) was reverse transcribed using random primers and AffinityScript reverse transcriptase (Stratagene). Reverse-transcription polymerase chain reaction (RT-PCR) used standard cycling conditions on an Applied Biosystems Veriti thermal cycler. Primary cell IL-15 quantitative PCR (qPCR) was performed using PerfeCTa SYBR Green FastMix, ROX (Quanta Biosciences). Primers were designed using Primer3 software. IL-15R qPCR used TaqMan primers and probes with Universal Mastermix (Applied Biosystems). Custom-designed TaqMan low-density arrays were run as previously described.28 Data were analyzed using RQ Manager 1.2.4 software (Applied Biosystems). The RT2 human tumor metastasis profiler arrays (catalog number PAHS-028ZC-2; SABiosciences) were run using SYBR green ROX qPCR Mastermix (SABiosciences) according to the manufacturer’s instructions. All qPCR reactions were performed using the Applied Biosystems 7500/7900HT Real-PCR System. PCR primer details and assay IDs are listed in supplemental Table 2.
Flow cytometry and Annexin V assays
CXCR3 and PSGL-1 antibodies were obtained from eBioscience. Intracellular IL-15 was detected using anti-human IL-15–phycoerythrin antibodies/isotype control (R&D Systems) following fixation and permeabilization using BD Cytofix/Cytoperm (BD Biosciences) according to the manufacturer’s instructions. Apoptosis was determined via the Annexin V Apoptosis Detection Kit FITC (eBioscience) according to the manufacturer’s instructions. Data were acquired on a MACSQuant flow cytometer (Miltenyi Biotec), and results were analyzed using FlowJo7.2.4 software (Tree Star).
Western blotting
Cells were lysed in RIPA buffer (Sigma-Aldrich) with protease/phosphatase cocktail inhibitor (1:100 dilution, Sigma-Aldrich), and 10 μg total protein (determined using a bicinchoninic acid protein assay kit; Thermo Scientific) was run on a 12% Bis-Tris gel (Invitrogen) and transferred via iBlot (Invitrogen) to nitrocellulose membrane. Blocking was performed in 10% nonfat milk (0.05% Tween/phosphate-buffered saline). All antibodies were purchased from New England Biolabs and used at 1:1000 dilutions unless otherwise stated. Chemiluminescence was detected using West Femto supersignal (Thermo Scientific).
SD-1 growth curves and inhibitors
SD-1 and Sup-B15 cells were seeded into 24-well flat-bottom plates at 1.5 × 105 cells per mL. Viable cells were counted using a hemocytometer with trypan blue dead-cell exclusion. Cells were exposed to IL-15Rα neutralizing antibody (nAb) (R&D Systems), isotype control antibody (R&D Systems), or rhIL-15. Small-molecule inhibitors of signaling pathways were diluted in dimethylsulfoxide (DMSO) and added to cells with or without addition of 25 ng/mL IL-15, and growth was compared with cells grown in the same percentage of DMSO (vehicle controls). Cell division was assessed using the CellTrace Violet proliferation kit (Invitrogen) using Colcemid-treated cells as a nondividing control.29
MTT assays
A total of 120 µL of 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTT; 5 mg/mL) (Sigma-Aldrich) in phosphate-buffered saline was added to each well of 24-well plates containing SD-1 cells and incubated for 4 hours at 37°C. Following centrifugation and removal of supernatant, 600 µL of extraction reagent (Fisher Scientific) was added per well and agitated for 15 minutes. A total of 100 µL solution per well was then transferred to a 96-well plate and absorbance measured in a sunrise absorbance reader (Tecan) at 570 nm (background wavelength = 660 nm). Analysis used MagellanCE6 software (Tecan).
Statistics
Parametric data were analyzed using Student unpaired t tests. Nonparametric data were analyzed using Mann-Whitney U (2 groups, unpaired), Kruskal-Wallis (>2 groups, unpaired), or Wilcoxon matched-pairs signed rank (>2 groups, paired) tests. A P value of ≤.05 was considered significant. All analysis was carried out using GraphPad Prism software (La Jolla, CA).
Results
Leukemic blasts express IL-15 and IL-15 receptors, and levels correlate with CNS infiltration in xenografts
RT-PCR was used to investigate expression of the 2 IL-15 isoforms and 3 IL-15 receptor subunits in pre-B ALL cell lines (SD-1, Sup-B15, REH, and SEM) and 19 primary pre-B leukemic diagnostic samples (for patient and cell line details, see supplemental Table 1). All expressed the cell-surface/secreted LSP-IL-15 isoform, but expression of the cytoplasmic SSP-IL-15 isoform was detected in only 6 out of 19 primary samples (Figure 1A and supplemental Table 1). In addition, all samples expressed IL-15Rα and the common γ chain and 14 out of 17 primary samples expressed IL-15Rβ (Figure 1A). This suggests that the majority of ALL cells have the potential to secrete and respond to IL-15 using autocrine, juxtacrine, or paracrine signaling loops.
IL-15 and IL-15 receptor expression by pre-B ALL cells and correlation with CNS infiltration. (A) RT-PCR analysis of IL-15 isoform and IL-15 receptor expression by cell lines and primary patient samples. Expected sizes in brackets; LSP-IL-15 (201 bp), SSP-IL-15 (320 bp), IL-15Rα (402 bp), IL-15/2Rβ (403 bp), IL-15/2Rγ (447 bp), GAPDH (housekeeping control) (115 bp). Representative examples are shown; individual results are listed in supplemental Table 1. (B) SYBR green relative quantification of IL-15 mRNA expression in primary patient samples using SD-1 as the calibrator (expression in patient samples reported as fold change relative to the level of expression in SD-1 cells, arbitrarily set at 1). (C) Histologic analysis of murine brains and spinal cord from xenografts. SD-1, Sup-B15, and REH hematoxylin and eosin–stained sections showing dark purple leukemic cells infiltrating the meninges (thick arrows) (top panel, left to right). All original magnification ×10; black scale bars represent 100 µm. Immunohistochemistry for human CD45 confirms the human origin of these infiltrating cells (bottom panel; isotype control shown in small inset panel). Original magnification ×40; black scale bars represent 100 µm. Section through spinal cord from SD-1 xenograft (bottom right); stars mark the sites of leukemic infiltration in the meningeal covering of the spinal cord. Original magnification ×20; black scale bars represent 100 µm. (D) Time to hind-limb paralysis (HLP) of cell-line xenografts (4-8 mice per cell line). (E-H) TaqMan qPCR analysis of IL-15 and all 3 components of the IL-15 receptor complex in xenografted cell lines. Three independent cultures were analyzed per cell line, and all results are expressed relative to the level in Sup-B15 cells, arbitrarily set at 1.0. All data are mean ± standard error of the mean and were analyzed by Student t tests comparing SD-1 cells to each of the other cell lines. ***P < .001, **P < .01, *P < .05, n.s = not significant. RQ, relative quantification.
IL-15 and IL-15 receptor expression by pre-B ALL cells and correlation with CNS infiltration. (A) RT-PCR analysis of IL-15 isoform and IL-15 receptor expression by cell lines and primary patient samples. Expected sizes in brackets; LSP-IL-15 (201 bp), SSP-IL-15 (320 bp), IL-15Rα (402 bp), IL-15/2Rβ (403 bp), IL-15/2Rγ (447 bp), GAPDH (housekeeping control) (115 bp). Representative examples are shown; individual results are listed in supplemental Table 1. (B) SYBR green relative quantification of IL-15 mRNA expression in primary patient samples using SD-1 as the calibrator (expression in patient samples reported as fold change relative to the level of expression in SD-1 cells, arbitrarily set at 1). (C) Histologic analysis of murine brains and spinal cord from xenografts. SD-1, Sup-B15, and REH hematoxylin and eosin–stained sections showing dark purple leukemic cells infiltrating the meninges (thick arrows) (top panel, left to right). All original magnification ×10; black scale bars represent 100 µm. Immunohistochemistry for human CD45 confirms the human origin of these infiltrating cells (bottom panel; isotype control shown in small inset panel). Original magnification ×40; black scale bars represent 100 µm. Section through spinal cord from SD-1 xenograft (bottom right); stars mark the sites of leukemic infiltration in the meningeal covering of the spinal cord. Original magnification ×20; black scale bars represent 100 µm. (D) Time to hind-limb paralysis (HLP) of cell-line xenografts (4-8 mice per cell line). (E-H) TaqMan qPCR analysis of IL-15 and all 3 components of the IL-15 receptor complex in xenografted cell lines. Three independent cultures were analyzed per cell line, and all results are expressed relative to the level in Sup-B15 cells, arbitrarily set at 1.0. All data are mean ± standard error of the mean and were analyzed by Student t tests comparing SD-1 cells to each of the other cell lines. ***P < .001, **P < .01, *P < .05, n.s = not significant. RQ, relative quantification.
qPCR indicates that primary ALL samples show a spectrum of IL-15 mRNA expression (Figure 1B). High expression has been shown to predict cytospin-positive CNS disease at diagnosis and the risk of CNS relapse.3 To examine the link with CNS disease further, we used a xenograft model to investigate CNS infiltration by pre-B cell lines. All cell lines tested were capable of CNS infiltration (Figure 1C) but with different kinetics as measured by time to HLP, a surrogate marker for leukemic infiltration of the leptomeninges (Figure 1D). Higher expression levels of mRNA encoding IL-15 and all 3 components of the IL-15 receptor (Figure 1E-H) were associated with shorter time to HLP (Figure 1D).
Pre-B ALL cells can respond directly to IL-15
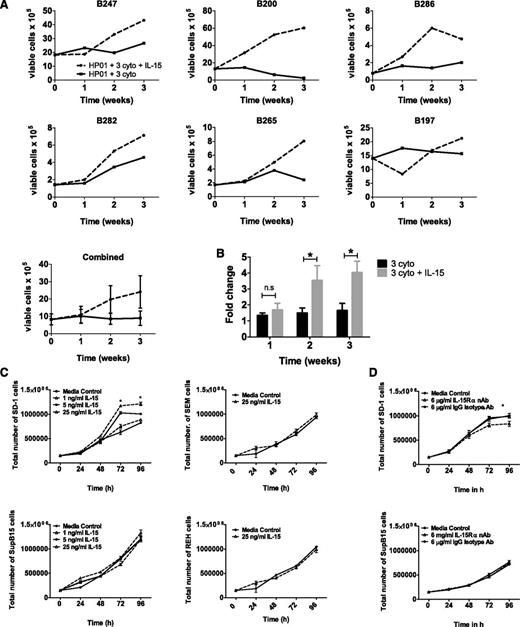
To test whether IL-15 mediates direct effects on pre-B ALL cells, 6 primary samples (patient details in supplemental Table 1) and 4 pre-B ALL cell lines (SD-1, Sup-B15, REH, and SEM) were exposed to IL-15 in suspension culture and the effects on growth were examined. A total of 5 out of 6 primary samples (Figure 2A) showed increased cell numbers in the presence of IL-15. To account for the wide variation in starting cell numbers and differing rates of proliferation between primary samples, we also calculated fold change in cell numbers compared with baseline (Figure 2B); this confirmed that IL-15 significantly enhanced cell growth when added to standard 3-cytokine mix. The effect of IL-15 on cell lines is more restricted and is shown in Figure 2C. SD-1 cells show a dose-dependent response to exogenous IL-15, but other cell lines appear to be IL-15 independent. To investigate whether endogenous IL-15 also plays a role, cells were incubated with an IL-15Rα nAb (in the absence of an exogenous source of IL-15) to block autocrine IL-15 signaling but leave signaling from IL-2 (which uses the same β/γ receptor subunit) and other cytokines (that use the common γ chain) unaffected. Exposure to IL-15Rα nAbs significantly decreased cell numbers at both 72 and 96 hours in SD-1 but not Sup-B15 cells (Figure 2D).
Pre-B ALL primary cells and SD-1 cells show increased growth in the presence of IL-15. (A) Six different primary pre-B ALL samples were grown in optimized 3-cytokine mix with or without the addition of IL-15 (25 ng/mL). Numbers of viable blast cells were measured at weeks 1, 2, and 3 by flow cytometry. Each sample is shown individually, and then results from all 6 experiments are pooled (bottom panel; data represent mean ± standard error of the mean). (B) Fold change in cell numbers compared with baseline (starting cell count at initiation of the experiment) was calculated for the 2 experimental conditions in each of the 6 primary samples. Data were analyzed using a Wilcoxon matched-pairs sign rank test, and bars display mean ± standard error of the mean. (C) Growth of SD-1, Sup-B15, REH, and SEM cells following treatment with 1, 5, or 25 ng/mL IL-15 for up to 96 hours. (D) Growth of SD-1 and SupB-15 cells following treatment with media alone, 6 μg/mL isotype control antibody, or 6 μg/ml IL-15Rα nAb. Viable cells were determined using trypan blue. Data represent mean ± standard error of the mean. Data were analyzed using an unpaired Student t test (*P < .05) and are representative of 3 independent experiments carried out per cell line.
Pre-B ALL primary cells and SD-1 cells show increased growth in the presence of IL-15. (A) Six different primary pre-B ALL samples were grown in optimized 3-cytokine mix with or without the addition of IL-15 (25 ng/mL). Numbers of viable blast cells were measured at weeks 1, 2, and 3 by flow cytometry. Each sample is shown individually, and then results from all 6 experiments are pooled (bottom panel; data represent mean ± standard error of the mean). (B) Fold change in cell numbers compared with baseline (starting cell count at initiation of the experiment) was calculated for the 2 experimental conditions in each of the 6 primary samples. Data were analyzed using a Wilcoxon matched-pairs sign rank test, and bars display mean ± standard error of the mean. (C) Growth of SD-1, Sup-B15, REH, and SEM cells following treatment with 1, 5, or 25 ng/mL IL-15 for up to 96 hours. (D) Growth of SD-1 and SupB-15 cells following treatment with media alone, 6 μg/mL isotype control antibody, or 6 μg/ml IL-15Rα nAb. Viable cells were determined using trypan blue. Data represent mean ± standard error of the mean. Data were analyzed using an unpaired Student t test (*P < .05) and are representative of 3 independent experiments carried out per cell line.
These experiments show that pre-B ALL samples can respond directly to the addition of IL-15 in culture, resulting in increased growth.
Mechanism of action of IL-15 in promoting pre-B ALL growth
To investigate the mechanism of IL-15 action on pre-B ALL blasts, Sup-B15 and SD-1 cells were studied further, because they carry the same leukemic translocation, t(9;22) BCR-ABL, but differ markedly in their IL-15 expression levels and speed of onset of CNS disease (high in SD-1 cells and low in Sup-B15 cells; Figure 1D).
IL-15 protein is difficult to detect due to its highly controlled secretion and fast turnover.25,30 Consistent with previous reports,18 IL-15 secretion was undetectable using an enzyme-linked immunosorbent assay (sensitivity ≥8 pg/mL; data not shown). However, intracellular IL-15 protein was detectable by flow cytometry in SD-1 cells, with lower levels in Sup-B15 (Figure 3A). Western blotting confirmed all 3 IL-15 receptor components are expressed by both cell lines, with higher levels of IL-15Rα in SD-1 than Sup-B15 (Figure 3B).
IL-15 has no effect on apoptosis in SD-1 cells. (A) Intracellular IL-15 protein levels in SD-1 and Sup-B15 cells, measured by flow cytometry with isotype- (shaded) and IL-15–specific antibodies (open). Corrected mean fluorescence intensity (MFI) represents MFI IL-15PE − MFI isotype control. (B) Levels of IL-15Rα, β, and γ in SD-1 and Sup-B15 cells were determined by western blot with β-tubulin as a loading control. MOLT-4 cells expressing high levels of IL-15Rα were used as a positive control. (C) Western blot for BCL-xL and BCL-2 protein expression in SD-1 cells treated with IL-15 (1, 5, and 25 ng/mL) for 72 hours. (D) PARP cleavage in SD-1 cells treated with IL-15 (25 ng/mL) for 96 hours. The positive-control lane contains SD-1 cells treated with the apoptosis-inducing agent AA2 (50 μM) for 1 hour. Histone H3 was used as loading control. (E) Annexin V-FITC and propidium iodide staining of SD-1 cells treated with (right panel) or without (central panel) 25 ng/mL IL-15 for 96 hours. The positive control shows cells exposed to AA2 (50 μM) for 1 hour (left). (F) SD-1 cells (dark bars) and the dexamethasone sensitive cell line CCRF-CEM (light bars; used as a positive control) were exposed to vehicle (0.1% EtOH) or dexamethasone (100 nM, 1 μM, or 10 μM) for 48 hours. Percentage apoptosis (sum of early and late apoptosis) of these cells was then measured using an Annexin V assay. Data represent mean ± standard error of the mean with n = 3 for each cell line. (G) SD-1 (dark bars) and CCRF-CEM cells (light bars) were exposed to similar conditions as panel C for 48 hours, and viability was assessed using an MTT assay. Data show viability (% of control) compared with untreated control. (H) MTT assay of SD-1 (dark bars) and CCRF-CEM (light bars) cells exposed to dexamethasone (Dex; 10 µM) ± IL-15Rα nAb/isotype control for 72 hours. In panels F to H, data represent mean ± standard error of the mean (n = 3) and were analyzed using a Kruskal Wallis test with Dunn’s multiple comparison test. **P < .01, ***P < .001. FCS, fetal calf serum; PE, phycoerythrin; PI, propidium iodide.
IL-15 has no effect on apoptosis in SD-1 cells. (A) Intracellular IL-15 protein levels in SD-1 and Sup-B15 cells, measured by flow cytometry with isotype- (shaded) and IL-15–specific antibodies (open). Corrected mean fluorescence intensity (MFI) represents MFI IL-15PE − MFI isotype control. (B) Levels of IL-15Rα, β, and γ in SD-1 and Sup-B15 cells were determined by western blot with β-tubulin as a loading control. MOLT-4 cells expressing high levels of IL-15Rα were used as a positive control. (C) Western blot for BCL-xL and BCL-2 protein expression in SD-1 cells treated with IL-15 (1, 5, and 25 ng/mL) for 72 hours. (D) PARP cleavage in SD-1 cells treated with IL-15 (25 ng/mL) for 96 hours. The positive-control lane contains SD-1 cells treated with the apoptosis-inducing agent AA2 (50 μM) for 1 hour. Histone H3 was used as loading control. (E) Annexin V-FITC and propidium iodide staining of SD-1 cells treated with (right panel) or without (central panel) 25 ng/mL IL-15 for 96 hours. The positive control shows cells exposed to AA2 (50 μM) for 1 hour (left). (F) SD-1 cells (dark bars) and the dexamethasone sensitive cell line CCRF-CEM (light bars; used as a positive control) were exposed to vehicle (0.1% EtOH) or dexamethasone (100 nM, 1 μM, or 10 μM) for 48 hours. Percentage apoptosis (sum of early and late apoptosis) of these cells was then measured using an Annexin V assay. Data represent mean ± standard error of the mean with n = 3 for each cell line. (G) SD-1 (dark bars) and CCRF-CEM cells (light bars) were exposed to similar conditions as panel C for 48 hours, and viability was assessed using an MTT assay. Data show viability (% of control) compared with untreated control. (H) MTT assay of SD-1 (dark bars) and CCRF-CEM (light bars) cells exposed to dexamethasone (Dex; 10 µM) ± IL-15Rα nAb/isotype control for 72 hours. In panels F to H, data represent mean ± standard error of the mean (n = 3) and were analyzed using a Kruskal Wallis test with Dunn’s multiple comparison test. **P < .01, ***P < .001. FCS, fetal calf serum; PE, phycoerythrin; PI, propidium iodide.
The observed increased cell numbers in the presence of IL-15 (Figure 2) could reflect reduced apoptosis or an ability to withstand growth arrest under conditions of serum depletion. IL-15 has powerful antiapoptotic effects in T and B cells.8,31 Therefore, we investigated whether IL-15 affected apoptosis in SD-1 cells. There was no alteration in BCL-xL and BCL-2 levels (Figure 3C) and no differences in apoptosis between IL-15 and untreated SD-1 cells measured by poly(ADP ribose) polymerase (PARP) cleavage (Figure 3D), Annexin V staining (Figure 3E), or caspase-3 cleavage (supplemental Figure 1a). To uncover a potential antiapoptotic role for IL-15 in the context of induced apoptotic stress, SD-1 cells were treated with dexamethasone (100 nM, 1 μM, and 10 μM) for 48 hours to induce apoptosis. Dexamethasone had no effect on SD-1 apoptosis or viability, as assessed using Annexin V (Figure 3F) and MTT (Figure 3G), respectively. Importantly, this could not be overcome by treatment with IL-15Rα nAb (Figure 3H), suggesting that IL-15-IL-15Rα signaling is not responsible for the resistance of SD-1 cells to dexamethasone-induced apoptosis.
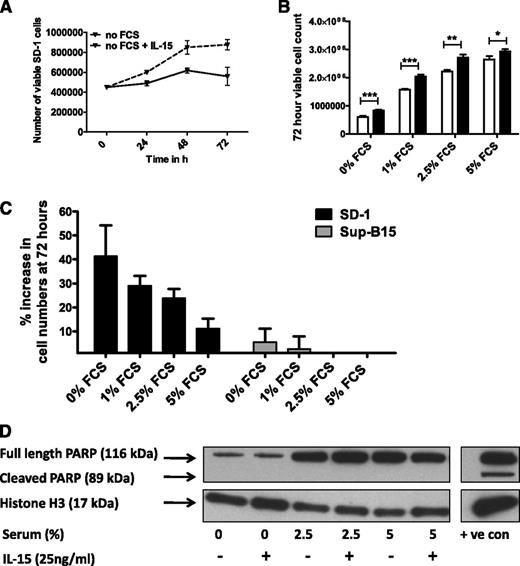
Together, these findings argue against reduced apoptosis being responsible for the higher plateau in IL-15–treated SD-1 cells. An alternative explanation is that IL-15 prevents growth arrest under conditions of serum depletion (as seen after 72-96 hours in culture without replenishment of growth media). To test this, SD-1 and Sup-B15 cells were grown under no/low-serum conditions with or without IL-15. IL-15 significantly enhanced the proliferation of SD-1 cells growing under conditions of no serum, 1% serum, and 2.5% serum (Figure 4A-B and supplemental Figure 1). Maximal benefit (in terms of percentage increase in growth with IL-15 treatment) was seen with 0% and 1% serum conditions (Figure 4B-C). Sup-B15 cells showed no response (Figure 4C). Again, there was no evidence that the IL-15 growth advantage was due to rescue from apoptosis (Figure 4D and supplemental Figure 1).
IL-15 promotes growth of SD-1 cells under low-serum conditions, which is independent of apoptosis. (A) SD-1 cells were grown in RPMI media without addition of fetal calf serum, with (dashed line) or without (solid line) 25 ng/mL IL-15. The growth curve from 1 representative experiment of 3 is shown. (B) SD-1 cells were grown in RPMI supplemented with increasing concentrations of fetal calf serum with (black bars) or without (white bars) 25 ng/mL of IL-15. The total number of viable cells in each group at 72 hours is shown. Data represent mean ± standard error of the mean from 3 independent experiments. Data were analyzed by a Mann-Whitney U test. *P < .05, **P < .01, ***P < .001. (C) Percentage increase in cell counts of IL-15–treated SD-1 (dark bars) and Sup-B15 (light bars) compared with media control. Data represent mean ± standard error of the mean from 3 independent experiments. SD-1 data are from the same experiments as in panel B above, represented in a different format. (D) PARP cleavage analysis of SD-1 cells exposed to increasing concentrations of serum ±25 ng/mL IL-15 for 96 hours. The positive-control lane represents SD-1 cells exposed to the apoptosis-inducing agent AA2 (50 μM) for 1 hour. Histone H3 was used as a loading control. FCS, fetal calf serum.
IL-15 promotes growth of SD-1 cells under low-serum conditions, which is independent of apoptosis. (A) SD-1 cells were grown in RPMI media without addition of fetal calf serum, with (dashed line) or without (solid line) 25 ng/mL IL-15. The growth curve from 1 representative experiment of 3 is shown. (B) SD-1 cells were grown in RPMI supplemented with increasing concentrations of fetal calf serum with (black bars) or without (white bars) 25 ng/mL of IL-15. The total number of viable cells in each group at 72 hours is shown. Data represent mean ± standard error of the mean from 3 independent experiments. Data were analyzed by a Mann-Whitney U test. *P < .05, **P < .01, ***P < .001. (C) Percentage increase in cell counts of IL-15–treated SD-1 (dark bars) and Sup-B15 (light bars) compared with media control. Data represent mean ± standard error of the mean from 3 independent experiments. SD-1 data are from the same experiments as in panel B above, represented in a different format. (D) PARP cleavage analysis of SD-1 cells exposed to increasing concentrations of serum ±25 ng/mL IL-15 for 96 hours. The positive-control lane represents SD-1 cells exposed to the apoptosis-inducing agent AA2 (50 μM) for 1 hour. Histone H3 was used as a loading control. FCS, fetal calf serum.
Thus, IL-15Rα neutralization reduces, and exogenous IL-15 stimulates, proliferation of SD-1 cells. These effects are not mediated via alterations in apoptosis. The significant growth advantage conferred by IL-15 treatment under low-serum conditions provides a potential explanation for the association between high IL-15 expression and leukemia relapse in the low-protein environment of the CNS.
IL-15 is not directly chemotactic but upregulates PSGL1, CXCR3, and SERPINE1
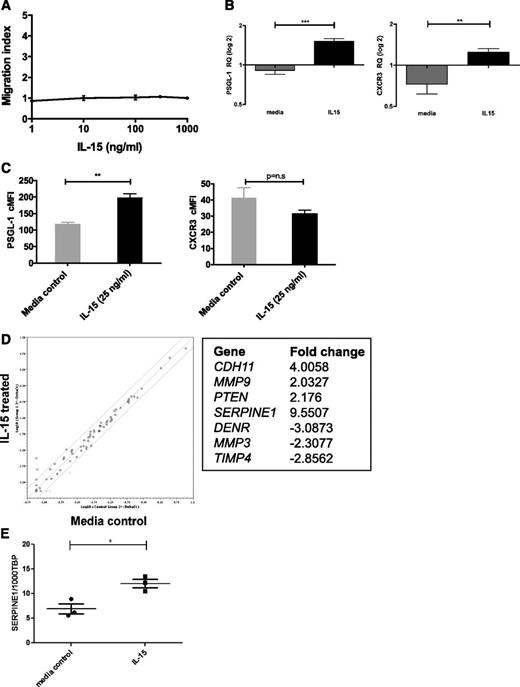
In addition to its growth effects, IL-15 could play a role in leukocyte trafficking, either acting as a chemoattractant32,33 or by modulating homing.34,35 Because IL-15 is expressed in the CNS,21,22 we tested whether IL-15 affected chemotaxis of SD-1 cells but were unable to show any migration toward an IL-15 gradient (Figure 5A). To investigate whether IL-15 indirectly regulates cellular migration or metastasis, 2 expression arrays were performed on IL-15–treated SD-1 cells. Firstly, a TaqMan low-density array of 30 human chemokine receptors, integrins, and selectins examined whether IL-15 treatment upregulated known leukocyte homing molecules. Only 2 out of 30 genes showed significant upregulation (supplemental Figure 2): the chemokine receptor CXCR3 and the selectin ligand platelet selectin glycoprotein ligand-1 (PSGL-1/SELPLG). Interestingly, both are implicated in blood–cerebrospinal fluid (CSF) barrier transit.36,37 These findings were confirmed by qPCR on an independent set of samples (Figure 5B), and upregulation of PSGL-1 was also seen by flow cytometry (Figure 5C). We went on to show that leukemic cells can migrate toward the CXCR3 ligand CXCL10 and that CXCL10 is detectable in CSF samples from patients with ALL. In addition, we show that the PSGL-1 receptor P-selectin is expressed on meningeal postcapillary venules in nonobese diabetic Cg-PrkdcscidIl2rgtm1Wjl/SzJ mice (supplemental Figure 3).
IL-15 is not directly chemotactic but induces expression of molecules associated with leukocyte trafficking. (A) SD-1 cells were placed in the top section of a bare filter transwell (5 μM pore size) and exposed to IL-15 (1, 10, 100, 300, or 1000 ng/mL) in the bottom section of the transwell. Transmigration was assessed following a 3-hour incubation period. The migration index was calculated by counting the total number of cells transmigrated in response to IL-15 as a proportion of the total number of cells that transmigrated in response to chemotaxis buffer alone (spontaneous migration). Data shown are from 1 experiment performed in triplicate. (B) SD-1 cells (n = 3 independent cultures) were treated with IL-15 (25 ng/mL) for 72 hours and levels of mRNA encoding PSGL-1 and CXCR3 measured by qPCR. Results are expressed as relative quantities with the level in media alone arbitrarily set to 1.0. (C) SD-1 cells (n = 3 independent cultures) were treated as in panel B, and the surface expression of PSGL-1 and CXCR3 was analyzed by fluorescence-activated cell sorter. Corrected mean fluorescence intensity (cMFI) represents MFI-specific antibody − MFI isotype control. (D) Following IL-15 treatment as in panel B above, expression of 95 genes associated with tumor metastasis was assessed using a commercially available RT2 PCR profiler array. Data are represented as a scatterplot (relative quantification log10), and solid lines represent twofold down- or upregulation. The 7 genes showing >twofold change with IL-15 treatment are listed. (E) Validation of hits from panel D by qPCR of SD-1 cells (n = 3 independent cultures) treated with media ±IL-15 (25 ng/ml) for 72 hours confirmed modest upregulation of Serpine1 by IL-15. Data show the number of copies of Serpine1 per 1000 copies of the housekeeping control gene TBP. All data are mean ± standard error of the mean and were analyzed by unpaired Student t tests. *P < .05, **P < .01, ***P < .001.
IL-15 is not directly chemotactic but induces expression of molecules associated with leukocyte trafficking. (A) SD-1 cells were placed in the top section of a bare filter transwell (5 μM pore size) and exposed to IL-15 (1, 10, 100, 300, or 1000 ng/mL) in the bottom section of the transwell. Transmigration was assessed following a 3-hour incubation period. The migration index was calculated by counting the total number of cells transmigrated in response to IL-15 as a proportion of the total number of cells that transmigrated in response to chemotaxis buffer alone (spontaneous migration). Data shown are from 1 experiment performed in triplicate. (B) SD-1 cells (n = 3 independent cultures) were treated with IL-15 (25 ng/mL) for 72 hours and levels of mRNA encoding PSGL-1 and CXCR3 measured by qPCR. Results are expressed as relative quantities with the level in media alone arbitrarily set to 1.0. (C) SD-1 cells (n = 3 independent cultures) were treated as in panel B, and the surface expression of PSGL-1 and CXCR3 was analyzed by fluorescence-activated cell sorter. Corrected mean fluorescence intensity (cMFI) represents MFI-specific antibody − MFI isotype control. (D) Following IL-15 treatment as in panel B above, expression of 95 genes associated with tumor metastasis was assessed using a commercially available RT2 PCR profiler array. Data are represented as a scatterplot (relative quantification log10), and solid lines represent twofold down- or upregulation. The 7 genes showing >twofold change with IL-15 treatment are listed. (E) Validation of hits from panel D by qPCR of SD-1 cells (n = 3 independent cultures) treated with media ±IL-15 (25 ng/ml) for 72 hours confirmed modest upregulation of Serpine1 by IL-15. Data show the number of copies of Serpine1 per 1000 copies of the housekeeping control gene TBP. All data are mean ± standard error of the mean and were analyzed by unpaired Student t tests. *P < .05, **P < .01, ***P < .001.
A second approach involved a RT2 PCR profiler array investigating 96 genes implicated in human tumor metastasis. IL-15 treatment had only a modest effect, with just 7 genes showing greater than twofold up- or downregulation (Figure 5D). Independent validation by qPCR confirmed that SERPINE1 was significantly, although modestly, upregulated by IL-15 treatment (Figure 5E and supplemental Figure 4).
Together, these experiments suggest that high IL-15 may increase interactions of circulating ALL cells with the blood-CSF and blood-LN barriers (via PSGL1/CXCR3) and/or promote invasiveness via SERPINE1 expression.
IL-15 induces STAT5 and ERK1/2 phosphorylation
Finally, we investigated downstream signaling pathways that might mediate the effects of IL-15 in ALL using phosphospecific antibodies for extracellular signal-regulated kinase 1/2 (ERK1/2), signal transducer and activator of transcription 5 (STAT5), IκBα, and Akt. Strong phosphorylation of STAT5 and ERK1/2 was seen within 15 minutes of the addition of IL-15 to SD-1 cells (Figure 6A-C). In addition, some activation of the nuclear factor κB (NF-κB) and phosphatidylinositol 3-kinase (PI3K)-Akt pathways is suggested by minor increases in phospho-IκBα and phospho-Akt, although these pathways are also constitutively activated in these cells (Figure 6A,D).
STAT5 and ERK1/2 signaling pathways are activated by IL-15 in SD-1 cells. (A) SD-1 cells were treated with IL-15 (25 ng/mL) for 15, 30, 45, 60, or 120 minutes. Cell lysates were analyzed by western blot using antibodies as indicated. (B) Western blot band density was analyzed via ImageJ software (National Institutes of Health) and adjusted densitometry values for phosphorylated STAT5 (pSTAT5) calculated by dividing the relative densities for pSTAT5 by the relative densities for total STAT5. (C-D) Adjusted densities values for (C) phosphorylated ERK1/2 (pERK1/2) and (D) phosphorylated IκBα (pIκBα) were calculated in a manner similar to above, except that relative densities for total ERK1/2 and IκBα were used, respectively.
STAT5 and ERK1/2 signaling pathways are activated by IL-15 in SD-1 cells. (A) SD-1 cells were treated with IL-15 (25 ng/mL) for 15, 30, 45, 60, or 120 minutes. Cell lysates were analyzed by western blot using antibodies as indicated. (B) Western blot band density was analyzed via ImageJ software (National Institutes of Health) and adjusted densitometry values for phosphorylated STAT5 (pSTAT5) calculated by dividing the relative densities for pSTAT5 by the relative densities for total STAT5. (C-D) Adjusted densities values for (C) phosphorylated ERK1/2 (pERK1/2) and (D) phosphorylated IκBα (pIκBα) were calculated in a manner similar to above, except that relative densities for total ERK1/2 and IκBα were used, respectively.
Given the potential therapeutic benefit of blocking IL-15 signaling in ALL, small-molecule inhibitors were investigated. Use of an ERK1/2 inhibitor (FR180204, Calbiochem) completely abolished the growth advantage conferred by IL-15 (Figure 7A). In contrast, STAT5 inhibition (573108, Calbiochem) reduced overall growth, although the growth advantage of exogenous IL-15 was still maintained (Figure 7B and supplemental Figure 5). A more potent mitogen-activated protein kinase kinase (MEK) inhibitor (U0126, Calbiochem), which lies upstream of ERK1/2 in the Raf/Ras/ERK pathway, profoundly reduced the growth of SD-1 cells and abolished any IL-15 stimulation of growth (Figure 7C and supplemental Figure 5). Both PI3K (LY 294002; Calbiochem) and NF-κB (IKK-2 inhibitor IV, Calbiochem) inhibitors had profound effects on leukemic cell growth both with and without IL-15 (Figure 7D), making assessment of their specific role in IL-15 signaling difficult.
IL-15–induced proliferation of SD-1 cells is mediated via the MEK/ERK signaling pathway. SD-1 cells were treated with (A) 10 μM ERK1/2 inhibitor (dashed lines), (B) 100 μM STAT5 inhibitor (dashed lines), (C) 10 μM MEK inhibitor (dashed lines), (D) 5 µM NF-κB inhibitor (IKK-2 inhibitor IV), or 20 µM PI3K inhibitor (LY 294002) or matched vehicle controls (0.1% DMSO) (solid lines) with (solid symbols) or without (open symbols) additional IL-15 (25 ng/mL). Viable cell numbers were determined using trypan blue. Data are mean ± standard error of the mean. Unpaired Student t tests were used to compare cell counts at 72 and 96 hours in drug-treated IL-15 vs no IL-15 groups. *P < .05, **P < .01, n.s = no significant difference. Vehicle-treated IL-15 vs no IL-15 groups showed a significant (P < .05) increase in cell numbers with IL-15 treatment at 72 and 96 hours as previously shown (Figure 2C). Representative graphs of 3 independent experiments are shown.
IL-15–induced proliferation of SD-1 cells is mediated via the MEK/ERK signaling pathway. SD-1 cells were treated with (A) 10 μM ERK1/2 inhibitor (dashed lines), (B) 100 μM STAT5 inhibitor (dashed lines), (C) 10 μM MEK inhibitor (dashed lines), (D) 5 µM NF-κB inhibitor (IKK-2 inhibitor IV), or 20 µM PI3K inhibitor (LY 294002) or matched vehicle controls (0.1% DMSO) (solid lines) with (solid symbols) or without (open symbols) additional IL-15 (25 ng/mL). Viable cell numbers were determined using trypan blue. Data are mean ± standard error of the mean. Unpaired Student t tests were used to compare cell counts at 72 and 96 hours in drug-treated IL-15 vs no IL-15 groups. *P < .05, **P < .01, n.s = no significant difference. Vehicle-treated IL-15 vs no IL-15 groups showed a significant (P < .05) increase in cell numbers with IL-15 treatment at 72 and 96 hours as previously shown (Figure 2C). Representative graphs of 3 independent experiments are shown.
Together, these results suggest that the Raf/Ras/ERK signaling pathway is directly involved in mediating IL-15 growth-promoting effects, whereas STAT5 does not seem to play a role in this pathway. PI3K and NF-κB may also be important either directly or indirectly in IL-15 action.
Discussion
Here, we have shown that IL-15 upregulates leukocyte trafficking molecules and promotes cell proliferation under normal and hostile conditions in pre-B ALL. Thus, we have established a mechanistic link between the effects of IL-15 on leukemic blasts and GWAS and microarray studies identifying IL-15 as a candidate gene for leukemia development,1 high-risk minimal residual disease,2 and CNS relapse.3
The GWAS data1,2 associate germline SNPs in IL-15 with leukemia and therefore cannot distinguish the production of IL-15 by leukemic blasts themselves or by the host microenvironment as the most important determinant of biological response. We show evidence for both; inhibition of autocrine or juxtacrine signaling via IL-15Rα neutralization reduced the growth of SD-1 cells, but larger growth-promoting effects were seen with exogenous IL-15, suggesting that paracrine signaling from the host microenvironment may be more important than autocrine signaling from the blasts themselves. All primary samples express IL-15, but a few samples lack expression of all components of the IL-15 receptor. This suggests that there may be heterogeneity between patients in the ability to respond to IL-15. Associations between IL-15 receptor expression and disease outcome have never been tested. Our sample size is too small to address this important question, but study of a larger cohort of patients could potentially identify patients that would benefit from blockade of IL-15 receptor signaling using pharmacologic inhibitors as discussed below.
High IL-15 expression in primary leukemic blasts is correlated with CNS disease.3 Leukemic deposits in the CNS grow within the leptomeninges bathed in CSF. CSF has a very low protein content,38 a condition under which we show the growth-enhancement effect of IL-15 is particularly marked. IL-15 can promote survival in serum-free conditions in NK cells,12 although in that case the effect was mediated via prevention of apoptosis rather than proliferation. We hypothesize that high levels of IL-15 facilitate engraftment and long-term survival in the CNS. IL-15 may be more important in the CNS microenvironment than in the bone marrow, where serum starvation is unlikely to be a major selective pressure. Importantly, IL-15 is produced in the CNS by glial cells,22 and high levels of IL-15 are associated with neuroinflammatory disorders such as multiple sclerosis,21 suggesting that IL-15 may be of particular importance in brain pathology.
In addition to local effects on cell proliferation and survival, IL-15 may facilitate extramedullary dissemination of ALL, promoting bulky mediastinal and LN disease4 and CNS relapse.3 IL-15 is directly chemotactic to T cells32,33 and NK cells39 and also indirectly influences leukocyte trafficking. IL-15Rα knockout mice have low numbers of circulating T and B cells and fewer leukocytes in lymph nodes, despite grossly normal B- and T-cell development.35 IL-15 also plays a part in the regulation of immature B-cell homing via modulation of interferon-γ–mediated homing pathways.34 The induction of PSGL-1 by IL-15 may alter leukocyte trafficking; PSGL-1 is a ligand for both L and P selectins, the major determinants of tethering and rolling in LN endothelial venules.40 In addition, P-selectin/PSGL-1 interactions are important determinants of cellular passage across the blood-CSF barrier.36 Our screen also identified the chemokine receptor CXCR3 and the protease SERPINE1 as inducible by IL-15. CXCR3 is highly expressed in CSF leukocytes, and its ligand CXCL10 is upregulated in the CSF of multiple sclerosis patients,41 suggesting that CXCR3 may be important in CNS cell entry. SERPINE1 is a serine protease inhibitor that inhibits apoptosis42 and plays important roles in cellular invasion and angiogenesis.43 It is associated with poor prognosis in a variety of cancers.44 Interestingly, the gene encoding SERPINE1 is a known ERK1/2 target.45
IL-15 induces phosphorylation of ERK1/2 and STAT5. Interestingly, both these signaling pathways are implicated in ALL pathogenesis. Constitutive activation of the Raf/Ras/MEK/ERK pathway is common in childhood ALL, and acquired Ras mutations are considered to be driver mutations.46 STAT5 activation is strongly associated with high-risk ALL subsets such as Philadelphia-positive ALL47 and “Philadelphia-like” ALL,48 both of which are associated with high-risk minimal residual disease and relapse. The growth advantage conferred by IL-15 is completely blocked by inhibition of ERK1/2. In contrast, STAT5 inhibition reduces overall leukemic cell growth but does not abolish the growth-promoting effects of IL-15, suggesting it may play a role in other, as-yet-unidentified mechanisms related to disease aggressiveness. The specific role of the PI3K/Akt pathways and NF-κB pathways needs further investigation. Our results show that inhibition of these pathways abrogates IL-15 growth effects, but due to the requirement of these pathways for basal leukemic cell growth in culture, further analysis will be required to determine whether these effects are direct or indirect.
Finally, it is worth noting that IL-15 may also influence the immune response to ALL. Administration of IL-15 has been suggested as a potential cancer treatment by boosting antitumor immune responses.49 Although our studies did not address the effects of IL-15 on host immune cells, our findings, along with others,18-20 suggest that in ALL the increased malignant-cell biological fitness associated with high IL-15 might outcompete an increased immune response. Whether IL-15 has similar effects on growth of nonhematologic malignancies is currently unknown, although IL-15 SNPs have also been linked to colon cancer.50
Importantly, this paper illustrates how high-throughput unbiased approaches such as GWAS and microarray data can lead to focused studies investigating key biological drivers of disease that can be exploited by drug therapy. Our study not only provides an insight into mechanisms by which IL-15 promotes leukemic cell growth and a propensity for LN and CNS involvement but also identifies potential drug targets.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Leukaemia and Lymphoma Research Childhood Leukaemia Cell Bank and all contributing centers and patients; E. Cosimo and A. Michie for help with cell-division analysis; V. Kelly, P. Diamanti, J. Moppett, M. Cummins, H. Blair, and J. Vormoor for help with primary samples; and O. Heidenreich and V. Saha for the supply of cell lines.
C.H. holds a Scottish Senior Clinical Fellowship (Scottish Funding Council). This work was supported by the Kay Kendall Leukaemia Fund (KKL454), Royal Hospital for Sick Children, Edinburgh Haematology Fund, and the Chief Scientist Office (SCD/08). G.J.G. holds an Medical Research Council programme grant (G0901113).
Authorship
Contribution: C.H., G.J.G., A.B., and R.C. designed the research and analyzed the data; C.H., M.T.S.W., Y.Y., C.C., A.B., and S.S. performed the research; K.E.C., R.M., A.T., A.S., and B.G. provided patient data and material; and all authors contributed to writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christina Halsey, Centre for Immunobiology, Institute of Infection, Immunity and Inflammation, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow G12 8QQ, United Kingdom; e-mail: chris.halsey@glasgow.ac.uk.