Key Points
The protein S SHBG-like domain and, more specifically, its LG1 subunit are important for binding and enhancement of TFPI.
TFPI binding to the protein S SHBG-like domain likely positions TFPI Kunitz domain 2 for optimal interaction with the active site of FXa.
Abstract
Protein S is a cofactor for tissue factor pathway inhibitor (TFPI), accelerating the inhibition of activated factor X (FXa). TFPI Kunitz domain 3 residue Glu226 is essential for enhancement of TFPI by protein S. To investigate the complementary functional interaction site on protein S, we screened 44 protein S point, composite or domain swap variants spanning the whole protein S molecule for their TFPI cofactor function using a thrombin generation assay. Of these variants, two protein S/growth arrest–specific 6 chimeras, with either the whole sex hormone–binding globulin (SHBG)-like domain (Val243-Ser635; chimera III) or the SHBG laminin G-type 1 subunit (Ser283-Val459; chimera I), respectively, substituted by the corresponding domain in growth arrest–specific 6, were unable to enhance TFPI. The importance of the protein S SHBG-like domain (and its laminin G-type 1 subunit) for binding and enhancement of TFPI was confirmed in FXa inhibition assays and using surface plasmon resonance. In addition, protein S bound to C4b binding protein showed greatly reduced enhancement of TFPI-mediated inhibition of FXa compared with free protein S. We show that binding of TFPI to the protein S SHBG-like domain enables TFPI to interact optimally with FXa on a phospholipid membrane.
Introduction
Tissue factor pathway inhibitor (TFPI) is a ∼41 kDa Kunitz-type protease inhibitor that consists of an acidic amino-terminal polypeptide, followed by 3 tandem Kunitz-type domains (Kunitz domains 1, 2, and 3) and a basic carboxy (C)-terminal tail.1 It circulates in plasma at a concentration of ∼1.6 nM.2,3 The majority (∼80%) of plasma TFPI is truncated and bound to lipoproteins, ∼5% is localized in storage granules within platelets, ∼5% circulates as free truncated variants, and only around 10% is considered to be free full-length TFPI,4 with this having the greatest anticoagulant activity.5-7
TFPI and its cofactor protein S downregulate tissue factor (TF)-induced thrombin generation, and a deficiency of either protein has been linked to an increased risk of venous thrombosis.8-10 TFPI specifically inhibits the initiation of coagulation through direct binding and inhibition of factor Xa (FXa) and, in a FXa-dependent manner, inhibition of the TF/factor VIIa (FVIIa) complex by forming a quarternary TF/FVIIa/FXa/TFPI complex.11,12 The P1 residue in the Kunitz domain 2 of TFPI is required for binding to FXa, whereas the P1 residue in Kunitz domain 1 is required for binding and inhibition of TF/FVIIa.13 The kinetic mechanism of FXa inhibition by TFPI is described as a 2-step process where TFPI first forms an immediate encounter complex with FXa (FXa/TFPI), followed by a slow isomerization into a final tight complex (FXa/TFPI*).14
Protein S, as well as being a well-established cofactor for activated protein C (APC), is a cofactor for TFPI and reduces the Ki for FXa inhibition by TFPI approximately tenfold.15 It thereby reduces the dissociation constant of the TFPI/FXa complex to close to the plasma concentration of free full-length TFPI (0.25 nM).15 The protein S enhancement of TFPI is dependent on the TFPI Kunitz domain 3, particularly TFPI Kunitz domain 3 residues Glu226 and Arg199.16,17 The complementary interaction site on protein S for TFPI has to date not been the subject of any report, and the mechanism behind this enhancement has yet to be fully defined.
Protein S is a 73-kDa, vitamin K–dependent, multidomain protein.9 It comprises an N-terminal γ-carboxylated glutamic acid (Gla) domain, a thrombin-sensitive region (TSR), 4 epidermal growth factor–like (EGF) domains and a C-terminal sex hormone–binding globulin (SHBG)-like domain.18 The SHBG-like domain is composed of 2 laminin G-type subunits, LG1 and LG2.19,20 Protein S circulates in plasma at concentrations of 20 to 25 µg/mL, with approximately 60% bound to C4b-binding protein (C4BP).21-23 The protein S/C4BP interaction is of high affinity (0.1 nM), and the binding site for C4BP is located in the protein S SHBG-like domain.24,25
In the present study, we investigated the functional interaction site of protein S for TFPI using a library of 44 protein S variants. Using single-point and composite variants, we examined the importance of solvent-exposed amino acid residues spanning the Gla-TSR-EGF1-EGF2-EGF3-EGF4 domains. The role of the protein S SHBG-like domain was investigated using 3 previously described protein S/growth arrest–specific 6 (Gas6) domain swap chimeras.24,25 Using thrombin generation and FXa inhibition assays, as well as a direct binding method, we demonstrate that the SHBG-like domain mediates the TFPI cofactor function of protein S.
Materials and methods
Generation and expression of protein S variants
Wild-type (WT) protein S and a panel of 34 protein S variants containing composite or single amino-acid substitutions spanning the Gla-TSR-EGF1-EGF2 domains have been generated and described previously.26,27 Only protein S Gla variants previously shown to bind normally to phospholipid membranes were evaluated for their TFPI cofactor function.27,28 In addition, the cDNA for 3 previously described protein S/Gas6 chimeras were available.24,25 In these, either the whole SHBG-like domain (Val243-Ser635; chimera III), or its individual LG1 (Ser283-Val459; chimera I) or LG2 (Ser460-Ser635; chimera II) subunits, were replaced by the corresponding domains of the homologous Gas6. Additional protein S mutants spanning the EGF1 and 3-4 domains were constructed through site-directed mutagenesis (KOD Hot Start DNA polymerase kit; Novagen) using the pcDNA/WT protein S vector as a template and mutagenic oligonucleotide primers (Thermo Scientific). Mutations were verified by DNA sequencing. The following mutations were introduced: N86A/E87Q/D88A/M91A (termed NEDM), K94A/T103A/K105A (termed KTK), D182N/E184Q/E186Q/E189Q (termed DEEE), R192Q/E201Q/D202N/D204N (termed REDD), D227N/K233Q (termed DK), K229Q/K230Q (termed KK), and D237N/K239Q/E242Q (termed DKE); see supplemental Table 1 on the Blood Web site for full details on substitutions in all 44 protein S variants.
Protein S purification and quantification
WT protein S and protein S variants were either concentrated in conditioned medium or purified using barium citrate precipitation29 followed by anion-exchange chromatography as previously described.27 Protein S concentrations of WT protein S and protein S single or composite variants (spanning Gla-TSR-EGF1-EGF2-EGF3-EGF4) were determined by a previously described in-house enzyme-linked immunosorbent assay (ELISA).26,27 The concentrations of purified WT protein S and protein S/Gas6 chimeras were determined by absorption at 280 nm using extinction coefficients (E1%, 1 cm) of 9.8, 10, 9.5, and 9.9 for WT protein S and chimeras I, II, and III, respectively. The extinction coefficients were predicted through the ProtParam Tool (ExPasy). Semiquantitative western blotting using a monoclonal antibody confirmed the validity of this approach (results not shown).
TFPI expression, purification, and quantification
TFPI was expressed, purified, and quantified as previously described.17 Briefly, the TFPI expression vector was transiently transfected into HEK293T cells. After a 3-day expression, full-length TFPI was purified in 2 steps; heparin Sepharose FF chromatography followed by affinity purification using an anti-TFPI Kunitz domain 1 antibody (Sanquin). The pure TFPI was quantified using an in-house ELISA. As described previously, TFPI expressed and purified according to this protocol is full-length and fully active and migrates as one band on a sodium dodecyl sulfate–polyacrylamide gel electrophoresis with an apparent molecular weight of 41 kDa.
Phospholipid vesicle preparation
Synthetic phospholipids (Avanti Polar Lipids) 1,2-Dioleoyl-sn-glycero-3-phosphoserine, 1,2-Dioleoyl-sn-glycero-3-phosphocholine, and 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine were mixed in a molar ratio of 20:60:20 and extruded as previously described.26
Thrombin generation assay by CAT
The TFPI cofactor function of protein S was assessed by calibrated automated thrombography (CAT) using a Fluoroscan Ascent FL plate reader (Thermo Labsystem) and Thrombinoscope software (Synapse BV), as described previously.26,27 Thrombin generation was initiated in 80 µL protein S–depleted plasma (Enzyme Research Laboratories) with 1 pM TF (Dade Innovin), 50 μM phospholipid vesicles, and 16.6 mM CaCl2 in a total volume of 120 µL. The amount of thrombin formed was monitored using 0.42 mM of a thrombin-sensitive fluorogenic substrate, Z-Gly-Gly-Arg-AMC (Bachem) in the presence or absence of TFPI (0-2 nM) and protein S (0-100 nM). Protein S was added either as concentrated and dialyzed conditioned media or as purified protein. The addition of concentrated conditioned media from mock-transfected HEK293 cells did not affect thrombin generation. To inhibit contact activation, corn trypsin inhibitor (Enzyme Research Laboratories) was added (30 µg/mL plasma in the screening experiments and 65 µg/mL plasma in protein S titrations). A polyclonal antibody against protein S (DAKO) and mixture of monoclonal antibodies against Kunitz domain 1, Kunitz domain 2, and C-terminus of TFPI (Sanquin) were used to show that any effects from the addition of protein S and TFPI were specific.
The TFPI cofactor function of protein S was initially assessed in 3 different protein S–depleted plasmas. One of the plasmas (frozen protein S–depleted plasma; Enzyme Research Laboratories) showed more reproducible results and was more sensitive to the addition of TFPI and protein S than the other two (lyophilized protein S–depleted plasma from Hyphen and Affinity Biologicals; results not shown) and was consequently used for all experiments reported in this study.
Binding of protein S to domain-specific monoclonal antibodies
Domain-specific monoclonal antibodies MK21 (Gla), MK41 (EGF3/EGF4), MK54 (EGF1), and MK67 (TSR),30 all at 1 µg/mL, were immobilized in a 96-well Maxisorp microplate in 50 mM sodium carbonate buffer, pH 9.6, at 4°C overnight. All subsequent incubations were carried out at room temperature, and wells were washed 3 times with 250 μL of 20 mM Tris-HCl pH 7.4, 150 mM NaCl, 3 mM CaCl2 (TBSCa2+), and 0.1% Tween 20 between each step. Wells were blocked with 200 μL 3% bovine serum antigen (BSA) in TBSCa2+ for 2 hours. Titrated protein S variants (0-10 nM for MK21, 0-15 nM for MK41 and MK54, and 0-1.5 nM for MK67) were incubated in the plate for 1 hour. Bound protein S was detected with a biotinylated monoclonal antiprotein S antibody (HTI), followed by a high sensitivity streptavidin-horseradish peroxidase diluted 1:10000 (Pierce).
Protein S-dependent APC-mediated FVa inactivation assay
Protein S enhancement of APC-mediated inactivation of a previously prepared and reported double mutant of FV, FV R506Q/R679Q,31 was assessed by determining the loss of FVa activity as described previously.26,27 Briefly, FV R506Q/R679Q was activated by human thrombin followed by the addition of hirudin. The activated FVa variant was used as a substrate for APC to determine the ability of protein S variants (0-100 nM) to enhance cleavage of Arg306 of FVa by 0.25 nM APC in the presence of phospholipids. After stopping the reaction by a dilution in ice-cold TBSCa2+ supplemented with 0.5% BSA, the remaining FVa activity was measured in a prothrombinase assay, as described previously.26
C4BP purification and characterization
Plasma-purified C4BP (Complement Technologies, Inc.), was further purified to remove bound protein S, essentially as previously described.32 Briefly, C4BP was incubated in 3 M guanidine and 20 mM Tris, pH 7.4, for 1 hour at room temperature and separated from protein S using a HiPrep 16/60 Sephacryl S-300 column (GE Healthcare). C4BP and protein S eluted in separate fractions, as expected. The C4BP pool was concentrated and the protein concentration was determined by absorption at 280 nm using an extinction coefficient (E1%, 1 cm) of 13.32.33 To assess the optimal C4BP/protein S ratio resulting in complete binding of protein S, different molar concentrations of C4BP and WT protein S were mixed in TBSCa2+ supplemented with 0.1% BSA and incubated for 1 hour at room temperature. The presence of free protein S was investigated using size-exclusion chromatography (Superose 12; GE Healthcare) followed by ELISA and western blotting. We found that when a molar ratio of 1.8:1 of C4BP and protein S were mixed, no free protein S was detected (results not shown).
FXa inhibition assay
FXa (0.5 nM; Enzyme Research Laboratories) activity was monitored by the cleavage of the chromogenic substrate S-2765 (Chromogenix), in the presence or absence of 2 nM TFPI and protein S variants (0-160 nM), in the presence of 25 μM phospohlipids and 5 mM CaCl2, as described previously.16,17 To study the enhancement of TFPI-mediated FXa inhibition by C4BP-bound protein S, WT protein S was preincubated with a twofold molar excess of C4BP in assay buffer for 1 hour at room temperature. TFPI cofactor function of C4BP-bound protein S was studied as described here previously. To derive the concentration of protein S required to reach 50% of the maximal enhancement of TFPI-mediated inhibition of FXa (EC50), the initial velocity (V0) of the S-2765 cleavage was plotted against the protein S concentration. The V0 was determined using nonlinear regression as previously described.15,16 The EC50 was determined by a one-phase exponential decay nonlinear curve fit.16,17
Binding of protein S to TFPI investigated by surface plasmon resonance
Results
Screening of protein S variants for the TFPI cofactor function in plasma
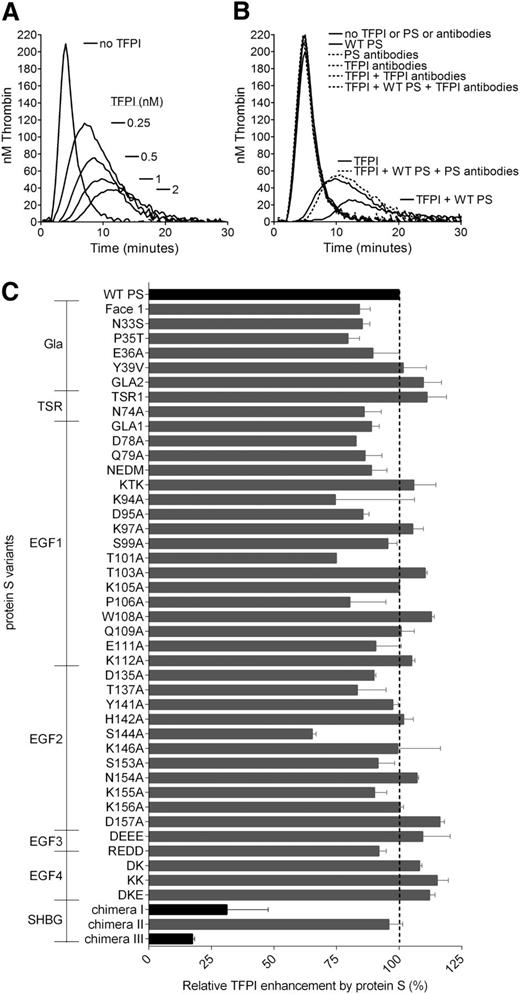
Thrombin generation was measured in protein S–depleted plasma. A dose-dependent decrease in thrombin generation was observed after the addition of increasing concentrations of TFPI (0-2 nM) in the absence of protein S. As expected, dose-dependent effects were detected by reduction of the peak and endogenous thrombin potential values, as well as an increase in lag time (Figure 1A). Inhibitory antibodies against TFPI did not have any effect on thrombin generation in the absence of added TFPI, suggesting that the anticoagulant pool of TFPI in the plasma was co-depleted with protein S (Figure 1B). This is consistent with the findings previously reported by Castoldi et al.34 The addition of WT protein S in the absence of TFPI had no effect on the thrombin generation, whereas it efficiently enhanced TFPI-mediated inhibition of thrombin generation, as reported previously.15,17,34 Inhibitory antibodies against TFPI and protein S were used to verify that inhibitory effects were a direct consequence of TFPI and TFPI cofactor function of protein S (Figure 1B).
Protein S enhancement of the TFPI-mediated inhibition of thrombin generation in protein S–depleted plasma. Thrombin generation was measured in protein S–depleted plasma supplemented with 50 µM phospholipids and 1 pM TF in the presence or absence of 0 to 2 nM (A) or 1 nM TFPI (B-C) and in the presence or absence of 50 nM WT PS (B-C). (A) A dose-dependent decrease in thrombin generation could be seen after the addition of increasing concentrations of TFPI. (B) Inhibitory antibodies against TFPI (a mix of 120 nM anti-TFPI Kunitz domain 1, 120 nM anti-TFPI Kunitz domain 2, and 120 nM anti-TFPI C-terminus) did not affect thrombin generation in protein S–depleted plasma, whereas they successfully repressed all inhibition of thrombin generation by TFPI in the presence or absence of protein S. In addition, inhibitory antibodies against protein S (2.8 µM; Dako) inhibited the enhancement of TFPI seen in the presence of protein S but had no effect on the absence of protein S. (C) The TFPI cofactor function of WT protein S and protein S variants was evaluated at 1 nM TFPI and 50 nM protein S. Pure WT protein S and WT protein S in concentrated conditioned media gave identical results. Protein S variants with amino acid substitutions in the Gla-TSR-EGF1-EGF2-EGF3-EGF4 domains (see supplemental Table 1 for full details of amino acid substitutions in the protein S variants used in this study) were screened in concentrated conditioned media. Protein S/Gas6 chimeras were purified before all experiments. All protein S variants were compared with WT protein S in concentrated conditioned media or were purified, as suitable. The relative protein S enhancement was determined as the decrease in peak height compared with WT protein S. The decrease in peak height in the presence of WT protein S compared with TFPI alone was set as 100% (n = 2). Results are expressed as mean ± standard deviation (SD). (A-B) Representative experiments are shown (n = 3). PS, protein S.
Protein S enhancement of the TFPI-mediated inhibition of thrombin generation in protein S–depleted plasma. Thrombin generation was measured in protein S–depleted plasma supplemented with 50 µM phospholipids and 1 pM TF in the presence or absence of 0 to 2 nM (A) or 1 nM TFPI (B-C) and in the presence or absence of 50 nM WT PS (B-C). (A) A dose-dependent decrease in thrombin generation could be seen after the addition of increasing concentrations of TFPI. (B) Inhibitory antibodies against TFPI (a mix of 120 nM anti-TFPI Kunitz domain 1, 120 nM anti-TFPI Kunitz domain 2, and 120 nM anti-TFPI C-terminus) did not affect thrombin generation in protein S–depleted plasma, whereas they successfully repressed all inhibition of thrombin generation by TFPI in the presence or absence of protein S. In addition, inhibitory antibodies against protein S (2.8 µM; Dako) inhibited the enhancement of TFPI seen in the presence of protein S but had no effect on the absence of protein S. (C) The TFPI cofactor function of WT protein S and protein S variants was evaluated at 1 nM TFPI and 50 nM protein S. Pure WT protein S and WT protein S in concentrated conditioned media gave identical results. Protein S variants with amino acid substitutions in the Gla-TSR-EGF1-EGF2-EGF3-EGF4 domains (see supplemental Table 1 for full details of amino acid substitutions in the protein S variants used in this study) were screened in concentrated conditioned media. Protein S/Gas6 chimeras were purified before all experiments. All protein S variants were compared with WT protein S in concentrated conditioned media or were purified, as suitable. The relative protein S enhancement was determined as the decrease in peak height compared with WT protein S. The decrease in peak height in the presence of WT protein S compared with TFPI alone was set as 100% (n = 2). Results are expressed as mean ± standard deviation (SD). (A-B) Representative experiments are shown (n = 3). PS, protein S.
The functional interaction site of protein S for TFPI was investigated using the library of 44 protein S variants. The screening of WT protein S and the protein S variants for their TFPI cofactor function was performed at 1 nM TFPI and 50 nM protein S (Figure 1C). The protein S Gla-TSR-EGF1-EGF2-EGF3-EGF4 variants were added as concentrated, conditioned media, whereas protein S/Gas6 chimeras were added as pure protein. WT protein S was studied both as concentrated conditioned media and pure protein; both enhanced TFPI equally well (results not shown). TFPI was efficiently enhanced by recombinant WT protein S. The great majority of protein S variants exhibited similar or only moderately decreased enhancement of TFPI compared with WT protein S (Figure 1C). Protein S enhancement of TFPI is known to be phospholipid dependent. To ensure that any decrease in TFPI cofactor function was not caused by a lack of binding to phospholipid membranes, we only studied protein S Gla variants already known to bind phospholipid membranes with the same affinity as WT protein S.27,28 All protein S Gla variants enhanced TFPI to a similar or moderately decreased level as WT protein S, suggesting that the protein S Gla domain has no other function in the enhancement of TFPI than binding to phospholipids. For 2 composite Gla-variants, Face1 and GLA2, covering the majority of Gla substitutions, this was also confirmed in FXa inhibition assays (results not shown). In contrast, we identified 2 variants with severely impaired TFPI cofactor function in these experiments (Figure 1C), protein S/Gas6 chimeras III and I, in which the whole SHBG-like domain (Val243-Ser635) or the LG1 subunit (Ser283-Val459), respectively, had been replaced by the corresponding domain in Gas6.
Protein S binding to domain-specific monoclonal antibodies and protein S enhancement of APC-mediated cleavage of FVa at Arg306
Binding of protein S/Gas6 chimeras I to III to domain-specific monoclonal antibodies was evaluated to assess the integrity of their domain structure. WT protein S and the protein S/Gas6 chimeras were titrated in a plate coated with monoclonal antibodies recognizing the Gla (MK21), TSR (MK67), EGF1 (MK54), or EGF3-EGF4 (MK41) domains. Bound protein was detected using a monoclonal antibody. Kd app values were obtained by fitting the binding curves to a one-site binding equation (Table 1). There was no significant difference (P > .05) in binding between the chimeras and WT protein S for any of the antibodies tested. The results suggest that replacement of the SHBG-like domain or its individual subunits did not lead to any major conformational changes within the remaining domains.
Furthermore, the ability of the protein S/Gas6 chimeras to enhance the APC-mediated cleavage of FVa at Arg306 was determined. For this, a FVa inactivation assay with R506Q/R679Q FVa was used. The FVa inactivation was performed in the presence of 0.25 nM APC and titrated (0-100 nM) WT protein S or protein S/Gas6 chimeras. After a 10-minute inactivation, the remaining FVa activity was measured in a prothrombinase assay. All chimeras efficiently enhanced the APC-mediated cleavage of Arg306 to the same extent as WT protein S (Table 1). The results suggest that the substitution of the SHBG-like domain or its individual subunits did not affect the enhancement of APC-mediated FVa cleavage at the Arg306 position. Importantly, these results also demonstrate that the chimeras bind phospholipids, because this is required for efficient enhancement of FVa inactivation in this assay.
Evaluation of the importance of the SHBG-like domain for the TFPI cofactor function of protein S in plasma
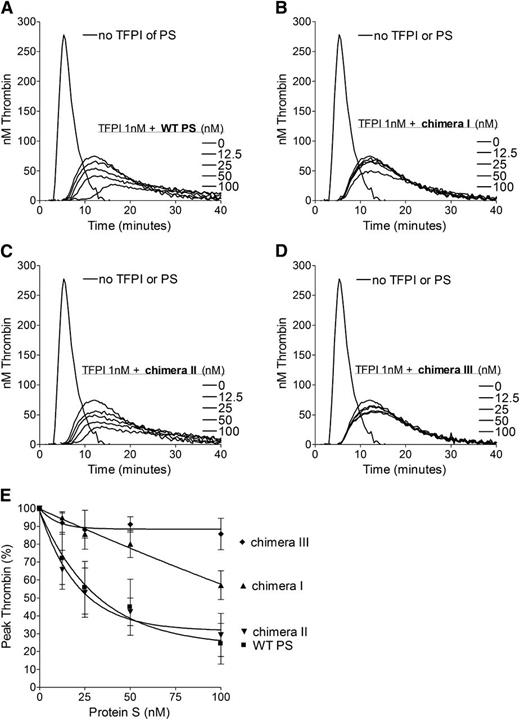
To further evaluate the importance of the SHBG-like domain in the TFPI cofactor function of protein S in plasma, pure WT protein S and protein S/Gas6 chimeras were titrated (0-100 nM) in the presence of 1 nM TFPI (Figure 2). Chimeras I and III both had minimal inhibitory effect on thrombin generation (Figure 2B,D), whereas chimera II enhanced TFPI to a similar extent as that of WT protein S (Figure 2A,C). When peak thrombin generation of each variant was compared (Figure 2E), chimera III was essentially unable to enhance TFPI. Chimera I reduced the peak height by only 45% at a concentration of 100 nM, whereas only 25 nM WT protein S and chimera II were needed for the same decrease in thrombin generation (Figure 2E). Under our experimental conditions, protein S had no effect on thrombin generation in the absence of TFPI (results not shown). These results suggest that the SHBG-like domain, and specifically the LG1 subunit of protein S (swapped in chimera I), is essential for TFPI cofactor function in plasma.
Enhancement of the TFPI-mediated inhibition of thrombin generation by WT protein S and protein S/Gas6 chimeras I to III. Thrombin generation was measured in protein S–depleted plasma supplemented with 50 μM phospholipids and 1 pM TF, in the presence or absence of 1 nM TFPI, and in the presence or absence of titrated (0-100 nM) WT protein S (A), chimera I (B), chimera II (C), or chimera III (D). Representative experiments are shown (n = 3). (E) Peak thrombin was plotted against protein S concentrations. The peak height in the presence of TFPI alone represents 100%. Results are expressed as mean ± SD (n = 3). PS, protein S.
Enhancement of the TFPI-mediated inhibition of thrombin generation by WT protein S and protein S/Gas6 chimeras I to III. Thrombin generation was measured in protein S–depleted plasma supplemented with 50 μM phospholipids and 1 pM TF, in the presence or absence of 1 nM TFPI, and in the presence or absence of titrated (0-100 nM) WT protein S (A), chimera I (B), chimera II (C), or chimera III (D). Representative experiments are shown (n = 3). (E) Peak thrombin was plotted against protein S concentrations. The peak height in the presence of TFPI alone represents 100%. Results are expressed as mean ± SD (n = 3). PS, protein S.
Protein S enhancement of TFPI-mediated FXa inhibition
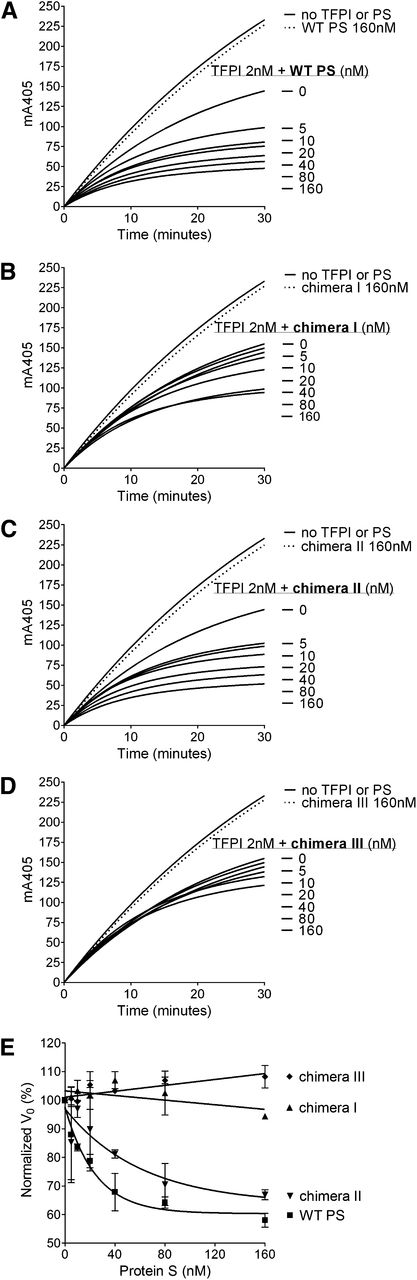
FXa inhibition assays with purified reagents were used to kinetically evaluate the enhancement of TFPI in its inhibition of FXa by WT protein S and protein S/Gas6 chimeras. Progress curves of FXa inhibition by TFPI and increasing concentrations of protein S are presented in Figure 3A-D. The initial rates of S-2765 hydrolysis by FXa in the presence of TFPI and increasing concentrations of protein S are plotted in Figure 3E, and the EC50 values have been derived. In agreement with the plasma-based assays, limited enhancement of TFPI was detected after the addition of chimera III (Figure 3D). A major decrease in the enhancement of TFPI by chimera I (Figure 3B) was also detected, whereas chimera II (Figure 3C) behaved similarly to WT protein S (Figure 3A) with only an approximately 2.7-fold increase in EC50 (39.6 ± 22.2 vs 14.6 ± 8.2 nM; presented as mean ± SD) (Figure 3E). These results confirm the importance of the protein S SHBG-like domain, and more specifically the LG1 subunit, in the TFPI cofactor function.
Enhancement of TFPI in the inhibition of FXa by WT protein S and protein S/Gas6 chimeras I to III. FXa activity (0.5 nM) was followed in real time through cleavage of S-2765 (200 µM) at 405 mm in the presence of 25 μM phospholipids, the presence or absence of TFPI (2 nM), and in increasing concentrations (0-160 nM) of WT protein S (A), chimera I (B), chimera II (C), or chimera III (D). Results from representative experiments are shown (n = 3). (E) The initial velocity (V0) was calculated and plotted against protein S concentration. Results are given as mean ± SD and are expressed as percentage of the V0 in the presence of TFPI alone (n = 3). PS, protein S.
Enhancement of TFPI in the inhibition of FXa by WT protein S and protein S/Gas6 chimeras I to III. FXa activity (0.5 nM) was followed in real time through cleavage of S-2765 (200 µM) at 405 mm in the presence of 25 μM phospholipids, the presence or absence of TFPI (2 nM), and in increasing concentrations (0-160 nM) of WT protein S (A), chimera I (B), chimera II (C), or chimera III (D). Results from representative experiments are shown (n = 3). (E) The initial velocity (V0) was calculated and plotted against protein S concentration. Results are given as mean ± SD and are expressed as percentage of the V0 in the presence of TFPI alone (n = 3). PS, protein S.
C4BP-bound protein S enhancement of TFPI-mediated FXa inhibition
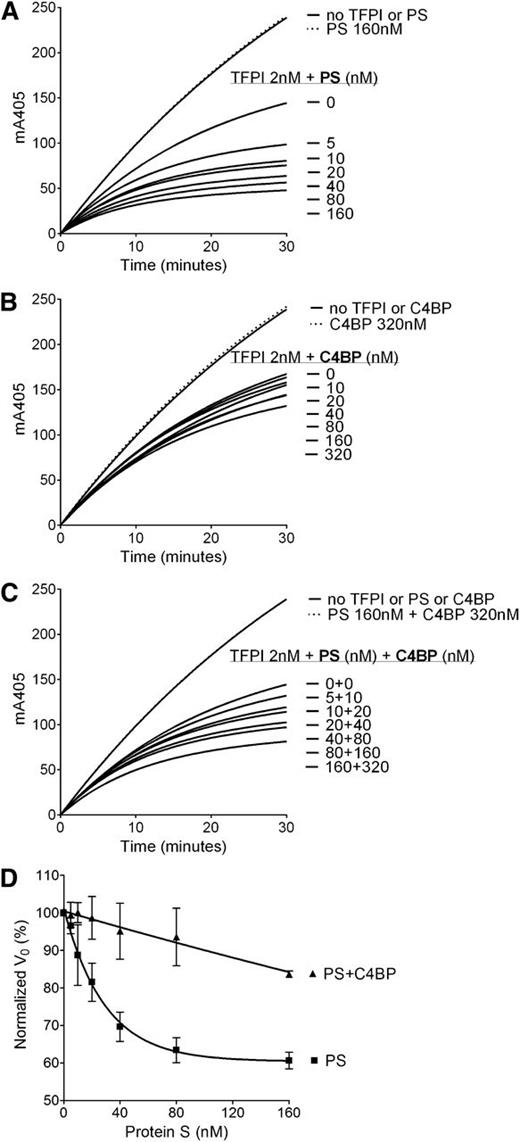
To further assess the role of the protein S SHBG-like domain in the TFPI cofactor function, the enhancement of TFPI-mediated inhibition of FXa by C4BP-bound WT protein S was studied and compared with the TFPI enhancement by free WT protein S using FXa inhibition assays (Figure 4). Although free WT protein S effectively enhanced TFPI-mediated inhibition of FXa in a dose-dependent manner (Figure 4A), a minimal enhancement by C4BP-bound WT protein S was detected (Figure 4C). These results support those of the protein S/Gas6 chimeras and strongly indicate the importance of protein S SHBG-like domain in its TFPI cofactor function.
Enhancement of TFPI inhibition of FXa by protein S–, C4BP-, and C4BP-bound protein S. FXa activity (0.5 nM) was followed in real time through cleavage of S-2765 (200 µM) at 405 mm in the presence of 25 μM phospholipids, the presence or absence of TFPI (2 nM), and in increasing concentrations (0-160 nM) of protein S (A), 0 to 320 nM C4BP (B), and C4BP-bound protein S in a 2:1 ratio (C). Representative experiments are shown (n = 3). (D) V0 was calculated and plotted against protein S concentration. Results are given as mean ± SD and are expressed as percentage of the V0 in the presence of TFPI alone (n = 3). PS, protein S.
Enhancement of TFPI inhibition of FXa by protein S–, C4BP-, and C4BP-bound protein S. FXa activity (0.5 nM) was followed in real time through cleavage of S-2765 (200 µM) at 405 mm in the presence of 25 μM phospholipids, the presence or absence of TFPI (2 nM), and in increasing concentrations (0-160 nM) of protein S (A), 0 to 320 nM C4BP (B), and C4BP-bound protein S in a 2:1 ratio (C). Representative experiments are shown (n = 3). (D) V0 was calculated and plotted against protein S concentration. Results are given as mean ± SD and are expressed as percentage of the V0 in the presence of TFPI alone (n = 3). PS, protein S.
Evaluation of binding of protein S to TFPI by SPR
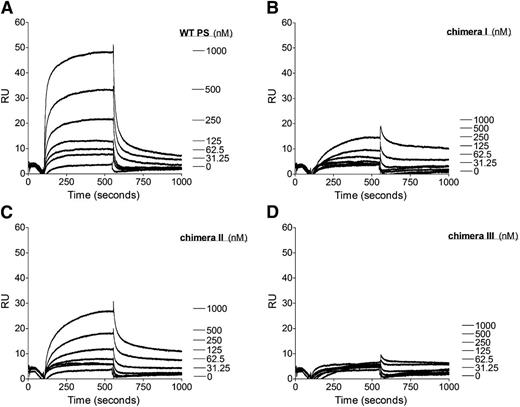
Our functional assays have shown that the protein S SHBG-like domain is essential for protein S to function as a cofactor for TFPI. SPR was performed to investigate the role of the protein S SHBG-like domain in the direct interaction with TFPI. Purified protein S (0-1000 nM) was perfused over immobilized TFPI. As previously shown, WT protein S bound to TFPI in a dose-dependent manner (Figure 5A).16,17,34 In agreement with previously published studies, no affinity for the TFPI/protein S interaction could be derived because a 2-phase association phase was observed and the binding curves did not reach equilibrium. Nevertheless, the results from the binding experiments correspond well with the results from the FXa inhibition assays. The substitution of the whole SHGB domain in chimera III resulted in an almost complete lack of binding interaction with TFPI (Figure 5D). Chimera I exhibited severely decreased binding to TFPI (Figure 5B), whereas chimera II interaction with TFPI was moderately decreased (Figure 5C). These results suggest that there is a direct interaction between the protein S SHBG-like domain and TFPI. Both LG subunits appear to be required for this interaction, but there is a more dominant role of LG1.
Binding of WT protein S and protein S/Gas6 chimeras I to III to TFPI studied with SPR. A CM5 chip was coupled with TFPI immobilized to 2500 response units. The flow cell was perfused with increasing concentrations (0-1000 nM) of WT protein S (A), protein S chimera I (B), protein S chimera II (C), or protein S chimera III (D). Results from a representative experiment are shown (n = 2). PS, protein S; RU, response units.
Binding of WT protein S and protein S/Gas6 chimeras I to III to TFPI studied with SPR. A CM5 chip was coupled with TFPI immobilized to 2500 response units. The flow cell was perfused with increasing concentrations (0-1000 nM) of WT protein S (A), protein S chimera I (B), protein S chimera II (C), or protein S chimera III (D). Results from a representative experiment are shown (n = 2). PS, protein S; RU, response units.
Discussion
The TFPI cofactor function of protein S plays an important role in the regulation of the initiation of coagulation. However, despite its importance, the molecular mechanism underlying the TFPI cofactor function of protein S has yet to be fully defined. In 2010, Ndonwi et al demonstrated the importance of the TFPI Kunitz domain 3 in the protein S cofactor function.16 We subsequently identified a functional interaction site for protein S at Glu226 within the TFPI Kunitz domain 3.17 In a recent study, Wood et al showed that the enhancement by protein S is limited to free full-length TFPI, either circulating in plasma or released from endothelial cells or platelets. In contrast, protein S does not enhance the activity of membrane-associated TFPI (TFPIβ).35 Because membrane-bound TFPI does not have a Kunitz domain 3, their study confirmed the importance of this domain for the protein S enhancement. Wood et al also created a TFPIβ variant containing a Kunitz domain 3 but, for reasons that are not clear, protein S was unable to enhance this TFPI variant.35
Although the protein S interaction site of TFPI is well characterized, the TFPI interaction site of protein S has not been determined. To explore this, we assessed the importance of solvent-exposed amino acid residues within the Gla-TSR-EGF1-EGF2-EGF3-EGF4 domains and the protein S SHBG-like domain, in the protein S enhancement of TFPI activity in thrombin generation assays. We identified 2 protein S/Gas6 chimeric variants showing substantial reductions in their TFPI cofactor function in plasma, a result that was confirmed in FXa inhibition assays. These findings suggest that the protein S SHBG-like domain (evaluated with chimera III) and mainly its LG1 subunit (evaluated with chimera I) are important for the TFPI cofactor function. The protein S/Gas6 chimeras have been characterized previously for their APC cofactor function and ability to bind C4BP.24,25,39 As previously reported, the whole SHBG swap, chimera III, has normal cofactor function for APC in its inactivation of FVa at residue Arg306, a result confirmed in our present study.24 This result is in agreement with results published by van Wijnen et al, who used a domain depletion approach and concluded that the SHBG-like domain is not important for the APC-mediated FVa cleavage.36 The 3 protein S/Gas6 chimeras also bind to domain-specific monoclonal antibodies with the same affinity as WT protein S. These results suggest that these domain swaps do not lead to any major conformational changes in Gla-TSR-EGF1-EGF2-EGF3-EGF4 domains.
That protein S can bind directly to TFPI has been established by SPR.16,17,34 In fact, the mechanism of protein S enhancement of TFPI depends on this direct interaction.17 Castoldi et al used immunoprecipitation experiments and suggested that free protein S and full-length TFPI form a complex in circulation.34 In the present study, we show that the protein S SHBG-like domain is directly involved in the protein S/TFPI interaction. The results obtained with the LG subunit chimeras suggest that both LG domains may contribute to the interaction. However, LG1 seems to be of greatest importance, because chimera II binds appreciably more than chimera I.
The protein S SHBG-like domain is known to contain the interaction site for C4BP. Both LG subunits are believed to be involved in the interaction with this protein, but despite numerous attempts, the interaction site has not been precisely identified.37-39 Hackeng et al have previously shown that the addition of a molar excess of C4BP to free protein S in normal plasma resulted in a 40% decrease of the TFPI enhancement by protein S in thrombin-generation assays.15 In addition, immunodepletion experiments showed that protein S bound to C4BP does not interact with TFPI.34 In the present study, we have compared the enhancement of TFPI-mediated FXa inhibition by C4BP-bound WT protein S to free protein S and demonstrated a dramatic decrease in the TFPI cofactor function. These results support a role of the protein S SHBG-like domain for the TFPI cofactor function as well as the direct interaction between protein S and TFPI. Interestingly, whereas both LG subunits have been shown to be important for the interaction with C4BP,37-39 our results suggests that the LG1 subunit of the SHBG-like domain may be of greater importance than the LG2 subunit for binding TFPI.
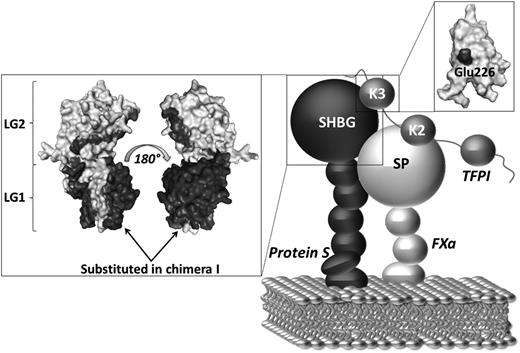
The affinity of TFPI for phospholipid membranes is very weak and is known to be dependent on the presence of the TFPI C-terminus.40,41 In contrast, it has also been shown that TFPI lacking the C-terminal tail can localize to phospholipid membranes in the presence of FXa, suggesting that direct binding of TFPI to phospholipids may only be important in limited physiologic contexts.40 In addition, protein S has been shown kinetically to enhance the initial interaction between TFPI and FXa. It has previously been hypothesized that protein S, through its high affinity for negatively charged phospholipids, brings TFPI in close proximity to FXa on an activated membrane and thereby increases the TFPI/FXa association rate.15-17,35 We propose that protein S and TFPI can associate in plasma through a direct interaction between the protein S SHBG-like domain and the TFPI Kunitz 3 domain, particularly with Glu226. Upon initiation of coagulation on phospholipid surfaces, protein S brings TFPI Kunitz domain 2 into proximity of the active site of the protease domain of FXa, thereby decreasing the concentration of TFPI needed for efficient inhibition of FXa (Figure 6).
Proposed mechanism of the protein S/TFPI/FXa complex assembly on a phospholipid surface. Our results suggest that the mechanism of protein S cofactor enhancement of TFPI involves formation of a complex between the SHBG-like domain of protein S and the Kunitz domain 3 (K3) of TFPI (particularly Glu226). Protein S helps localize TFPI onto an activated membrane surface in a position favorable for the interaction of Kunitz domain 2 (K2) with the serine protease domain (SP) of FXa. The boxes show the respective models of the protein S SHBG-like domain and the TFPI Kunitz domain 3. TFPI Glu226 and the residues substituted in protein S/Gas6 chimera I are highlighted in black. The TFPI Kunitz domain 3 and the protein S SHGB -domain models are adapted from Mine et al and Villoutreix et al, respectively.37,42
Proposed mechanism of the protein S/TFPI/FXa complex assembly on a phospholipid surface. Our results suggest that the mechanism of protein S cofactor enhancement of TFPI involves formation of a complex between the SHBG-like domain of protein S and the Kunitz domain 3 (K3) of TFPI (particularly Glu226). Protein S helps localize TFPI onto an activated membrane surface in a position favorable for the interaction of Kunitz domain 2 (K2) with the serine protease domain (SP) of FXa. The boxes show the respective models of the protein S SHBG-like domain and the TFPI Kunitz domain 3. TFPI Glu226 and the residues substituted in protein S/Gas6 chimera I are highlighted in black. The TFPI Kunitz domain 3 and the protein S SHGB -domain models are adapted from Mine et al and Villoutreix et al, respectively.37,42
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Dr Tsutomu Hamuro for kindly providing recombinant TFPI.
This work was supported by the British Heart Foundation (FS/11/60/28940).
Authorship
Contribution: N.R.-M., J.T.B.C., D.A.L., and J.A. designed the research, analyzed the results, and wrote the paper; N.R.M., H.M.A., and J.A. performed the experiments; and S.M.R. and B.D. contributed with essential reagents.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Josefin Ahnström, Imperial College London, Centre for Haematology, 5th Floor Commonwealth Building, Hammersmith Hospital Campus, Du Cane Rd, London, W12 0NN, United Kingdom; e-mail: j.ahnstrom@imperial.ac.uk.