Key Points
RAG1/2 and casein kinase 1 ε are key effectors of Sox4 function in progenitor B cells.
Sox4 induces B-cell differentiation by suppressing Wnt/β-catenin signaling and activating immunoglobulin gene recombination.
Abstract
Commitment of hematopoietic stem cells to B lineage precursors and subsequent development of B lineage precursors into mature B cells is stringently controlled by stage-specific transcription factors. In this study, we used integrated genetic approaches and systematically determined the role of Sry-related high mobility group box (Sox) 4 and the underlying molecular mechanisms in early B-cell development. We found that Sox4 coordinates multilevel controls in the differentiation of early stage B cells. At the molecular level, Sox4 orchestrates a unique gene regulatory program, and its function was predominantly mediated through a conventional Sox4-binding motif as well as an unconventional GA-binding protein α chain binding motif. Our integrated gene network and functional analysis indicated that Sox4 functions as a bimodular transcription factor and ensures B lineage precursor differentiation through 2 distinct mechanisms. It positively induces gene rearrangements at immunoglobulin heavy chain gene loci by transcriptionally activating the Rag1 and Rag2 genes and negatively regulates Wnt signaling, which is critical for self-renewal, by inducing the expression of casein kinase 1 ε. Our findings illustrate that Sox4 mediates critical fine-tuning of the 2 opposing forces in early B-cell development and also set forth a model for characterization of critical genes whose deficiency, like Sox4 deficiency, is detrimental to this process.
Introduction
Members of the Sry-related high mobility group box (Sox) transcriptional regulator family control critical developmental processes in embryonic and adult stages.1 Findings from our previous study and others demonstrated that Sox4 plays an indispensable role in B-cell development.2,3 However, the molecular mechanisms underlying the role of Sox4 as a transcriptional regulator in B lymphopoiesis remain obscure.
B-cell development from hematopoietic stem cells (HSCs) is a precisely regulated process whereby HSCs transit through distinct developmental stages to become mature B cells.4-6 Each stage is defined by expression of specific cell membrane receptors, signal transducers, and transactivators. Coordination among these different signaling components allows the cells to exit one stage and enter a subsequent stage. For instance, signaling pathways mediated by Wnt/β-catenin, Sonic hedgehog, and Notch are critical in HSCs and multipotent progenitors, and so is the signaling pathway of interleukin 7 (IL-7) in pro-B cells and that of pre-B receptor in pre-B cells.7-11 Although the mediators that execute these signaling cascades have been identified, the regulators that precisely switch the cascades on and off are yet to be defined.
Rag1 and Rag2 genes function nonredundantly in mediating variable, diverse, joining (VDJ) gene rearrangements at the immunoglobulin heavy chain (Igh) gene locus in pro-B cells.12,13 The expression of Rag1 and Rag2 is initiated in common lymphocyte progenitors and is critical in pro-B cells, as only pro-B cells with productively rearranged VDJ genes are able to transit to the next stage, pre-B. In Rag1−/− or Rag2−/− mice, B-cell development is arrested at the pro-B stage.14-16 Transcription factors Foxo1, Foxp1, Ebf1, E2A, Pax5, and Ikaros are involved in controlling the expression of Rag1 and Rag2.17-24 Conditional inactivation or complete ablation of several transcriptional regulators of Rag1 and Rag2 results in similar phenotypic defects. Although Sox4 deficiency induces a phenotype that partly resembles that induced by a Rag1 and Rag2 deficiency, it is not clear whether Sox4 is a transcriptional regulator of Rag1 and Rag2.
Wnt/β-catenin signaling is involved in long-term maintenance of HSCs in the bone marrow.7,25-27 However, when the strength of Wnt signaling in HSCs is increased through expression of a constitutively activated form of β-catenin, B lymphopoiesis as well as erythroid and myeloid development are severely impaired.28,29 In contrast, conditional inactivation of β-catenin in the hematopoietic system does not impede B-cell development.30,31 These findings imply that Wnt/β-catenin signaling is critical in HSCs and uncommitted or lineage-committed progenitors, but downregulation of Wnt/β-catenin signaling might be essential at a particular stage for normal development to progress.32 Little is known about the negative regulators of Wnt/β-catenin signaling in B-cell development.
Here we report our findings on the role of Sox4 in early B lymphopoiesis, obtained by integrated genetic approaches using a self-limiting in vitro conditional inactivation system. We demonstrate that Sox4 controls 2 obligatory processes required for B-cell development. First, Sox4 induces the expression of Rag1 and Rag2 and consequently controls recombination of VDJ genes at the Igh gene locus in pro-B cells. Second, Sox4 regulates the expression of casein kinase 1 ε (CK1ε) and ensures efficient downregulation of Wnt/β-catenin signaling. Our research has uncovered the novel mechanistic determinants of Sox4 function in early B-cell development.
Methods
Mice
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee of The University of Texas MD Anderson Cancer Center. Generation of loxp-flanked Sox4 allele mice,33 Vav-Cre mice,34 and Rosa26eYFP mice (Jackson Laboratory) was described previously.
OP9 and pro-B-cell coculture
Conditions used for coculturing OP9 and pro-B cells are described in supplemental Methods (available on the Blood Web site).
Viral vectors, transduction, and reporter analysis
Details of the viral vector construction, virus transduction, and reporter gene analysis are described in supplemental Methods.
Flow cytometry
Cell sorting and analysis was performed on Influx cell sorter and FACSCanto II or LSRFortessa flow cytometer (BD Biosciences), respectively. Details of the antibodies used in flow cytometry are provided in supplemental Methods.
Gene expression microarray analysis
Gene expression profiling was performed according to the manufacturer’s protocol (Ambion). Details of the analysis are provided in supplemental Methods.
bioChIP and ChIP-Seq data analysis
Biotin-mediated chromatin immunoprecipitation (bioChIP) was performed as described previously.35 Details of the chromatin immunoprecipitation sequencing (ChIP-Seq) data analysis are provided in supplemental Methods.
VDJ recombination analysis
Results
Establishment of a novel strategy to systematically study the molecular function of Sox4
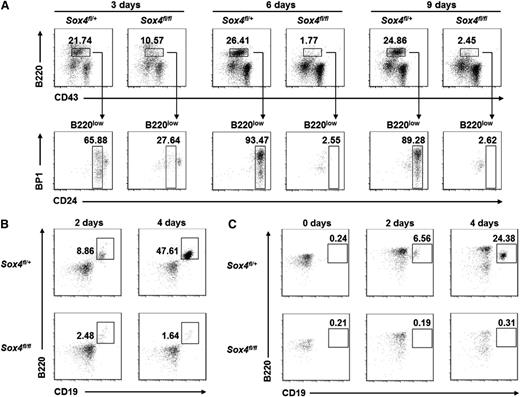
To systematically investigate the molecular function of Sox4, we at first attempted to establish pro-B-cell cultures from 14.5-day-old mouse fetal liver cells.37 As shown in Figure 1A, fetal liver cells from Sox4fl/+Vav-Cre mice were able to yield pro-B cells (B220lowCD24+) within 3 days. However, cells from Sox4fl/flVav-Cre mice failed to yield pro-B cells even after 9 days. Similarly, adult bone marrow cells from Sox4fl/+Vav-Cre mice could be cultured to generate pro-B cells, but those from Sox4fl/flVav-Cre mice could not (Figure 1B). Moreover, pre-pro-B cells sorted from Sox4fl/+Vav-Cre adult bone marrow could differentiate into pro-B cells (B220+CD19+) as early as day 2 in culture and significantly expanded after 4 days (Figure 1C), whereas those from Sox4fl/flVav-Cre mice failed to yield pro-B cells. These results established the important role of Sox4 in the differentiation of pre-pro-B cells to pro-B cells and demonstrated the necessity for a new strategy to yield sufficient numbers of Sox4-deficient cells to study the function of Sox4.
Failure of in vitro expansion of pro-B cells from Sox4 conditional knockout mice elucidates the essential role of Sox4 in early B-cell development. Cells were cultured in the presence of OP9 stromal cells and IL-7 and analyzed by flow cytometry. (A) Analysis of cultured embryonic fetal liver cells from Sox4fl/+Vav-Cre or Sox4fl/flVav-Cre mice for the presence of B220lowCD24+BP1− (fraction B) pro-B cells and B220lowCD24+BP1+ (fraction C) pro-B cells. (B) Analysis of cultured total bone marrow cells from Sox4fl/+Vav-Cre or Sox4fl/flVav-Cre mice for the presence of B220+CD19+ B cells. (C) Analysis of cultured pre-pro-B cells sorted from the bone marrow of Sox4fl/+Vav-Cre or Sox4fl/flVav-Cre mice for the presence of B220+CD19+ B cells. Hardy’s nomenclature4 is used, for example, pre-pro-B (fraction A), pro-B (fractions B and C), pre-B (fraction D), immature B (fraction E), and mature B (fraction F). Numbers indicate percentages corresponding to the cells outlined. All data are representative of 3 independent experiments.
Failure of in vitro expansion of pro-B cells from Sox4 conditional knockout mice elucidates the essential role of Sox4 in early B-cell development. Cells were cultured in the presence of OP9 stromal cells and IL-7 and analyzed by flow cytometry. (A) Analysis of cultured embryonic fetal liver cells from Sox4fl/+Vav-Cre or Sox4fl/flVav-Cre mice for the presence of B220lowCD24+BP1− (fraction B) pro-B cells and B220lowCD24+BP1+ (fraction C) pro-B cells. (B) Analysis of cultured total bone marrow cells from Sox4fl/+Vav-Cre or Sox4fl/flVav-Cre mice for the presence of B220+CD19+ B cells. (C) Analysis of cultured pre-pro-B cells sorted from the bone marrow of Sox4fl/+Vav-Cre or Sox4fl/flVav-Cre mice for the presence of B220+CD19+ B cells. Hardy’s nomenclature4 is used, for example, pre-pro-B (fraction A), pro-B (fractions B and C), pre-B (fraction D), immature B (fraction E), and mature B (fraction F). Numbers indicate percentages corresponding to the cells outlined. All data are representative of 3 independent experiments.
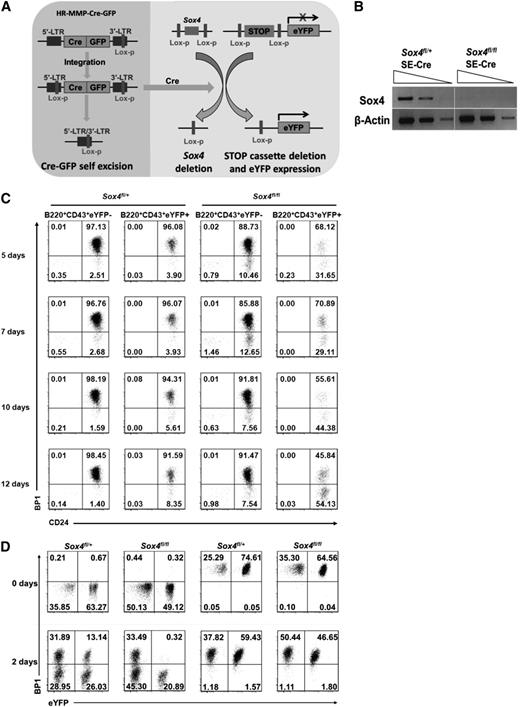
To this end, we expanded fetal liver pro-B cells from Sox4fl/fl;Rosa26eYFP mice and then induced Sox4 deletion by transducing the cells with Cre-expressing retrovirus. We found that pro-B cells with continuously expressed Cre readily underwent cell death (data not shown). To overcome this problem, we used a self-excising Cre (SE-Cre) vector that would express Cre to delete (1) the floxed Sox4 gene, (2) the stop cassette upstream of the enhanced yellow fluorescent protein (eYFP)–encoding sequence (so that eYFP was expressed as an indicator of Cre activity), and (3) the floxed Cre-encoding sequence (Figure 2A).38 Deletion of the Cre-encoding sequence would prevent further Cre expression and avoid Cre toxicity. With this method, we successfully established cultures of pro-B cells in which the floxed Sox4 gene was deleted (Figure 2B).
The new in vitro Sox4-knockout system uncovers the requirement of Sox4 at the transition from fraction B to fraction C in pro-B cells. (A) Schematic illustration of the deletion of floxed Sox4 by the SE-Cre recombinase. Cre also induced deletion of the stop cassette in eYFP reporter (to allow eYFP expression as an indicator of Cre activity) as well as deletion of its own gene to terminate its expression. (B) Semiquantitative reverse transcription–PCR (RT-PCR) analysis of Sox4 expression in fivefold serial dilutions of complementary DNA (cDNA) from eYFP+Sox4fl/+SE-Cre and eYFP+Sox4fl/flSE-Cre pro-B cells 10 days after Cre-induced floxed Sox4 deletion. β-Actin served as a loading control. (C) Flow cytometry analysis of cultured pro-B cells in the presence of IL-7 and OP9 stromal cells for the indicated numbers of days after the introduction of SE-Cre. In eYFP+Sox4fl/+SE-Cre and eYFP+Sox4fl/flSE-Cre cells, 1 and 2 Sox4 alleles were deleted, respectively. eYFP− cells served as an internal control. Note that the ratio of fraction B (CD24+BP1−) to fraction C (CD24+BP1+) cells was gradually increased in the eYFP+Sox4fl/flSE-Cre cultures. (D) Effect of Sox4 deletion on the differentiation of fraction B into fraction C. Fraction B and fraction C cells were each sorted out 9 days after the introduction of SE-Cre (day 0) and cultured for an additional 2 days before flow cytometry analysis. The percentages of cells in each quadrant are indicated (C-D). Data are representative of 2 (C) or 3 (B,D) independent experiments.
The new in vitro Sox4-knockout system uncovers the requirement of Sox4 at the transition from fraction B to fraction C in pro-B cells. (A) Schematic illustration of the deletion of floxed Sox4 by the SE-Cre recombinase. Cre also induced deletion of the stop cassette in eYFP reporter (to allow eYFP expression as an indicator of Cre activity) as well as deletion of its own gene to terminate its expression. (B) Semiquantitative reverse transcription–PCR (RT-PCR) analysis of Sox4 expression in fivefold serial dilutions of complementary DNA (cDNA) from eYFP+Sox4fl/+SE-Cre and eYFP+Sox4fl/flSE-Cre pro-B cells 10 days after Cre-induced floxed Sox4 deletion. β-Actin served as a loading control. (C) Flow cytometry analysis of cultured pro-B cells in the presence of IL-7 and OP9 stromal cells for the indicated numbers of days after the introduction of SE-Cre. In eYFP+Sox4fl/+SE-Cre and eYFP+Sox4fl/flSE-Cre cells, 1 and 2 Sox4 alleles were deleted, respectively. eYFP− cells served as an internal control. Note that the ratio of fraction B (CD24+BP1−) to fraction C (CD24+BP1+) cells was gradually increased in the eYFP+Sox4fl/flSE-Cre cultures. (D) Effect of Sox4 deletion on the differentiation of fraction B into fraction C. Fraction B and fraction C cells were each sorted out 9 days after the introduction of SE-Cre (day 0) and cultured for an additional 2 days before flow cytometry analysis. The percentages of cells in each quadrant are indicated (C-D). Data are representative of 2 (C) or 3 (B,D) independent experiments.
Within 10 days after the SE-Cre introduction, a relative increase in Sox4-deleted pro-B fraction B cells (B220+CD43+ CD24+BP1−) and a relative reduction in the pro-B fraction C cells (B220+CD43+CD24+BP1+) were consistently observed (Figure 2C). We reasoned that this change could have been caused by lack of differentiation from fraction B to fraction C in the absence of Sox4. We then sorted fraction B and C cells 9 days post-SE-Cre transduction and separately cultured them for 2 days (Figure 2D). eYFP− (ie, without the Sox4 gene deletion) fraction B cells from Sox4fl/+;Rosa26eYFP and Sox4fl/fl;Rosa26eYFP mice were able to differentiate into fraction C cells in 2 days. eYFP+ fraction B cells from Sox4fl/+;Rosa26eYFP mice (ie, with 1 copy of the Sox4 gene deleted) were able to differentiate into fraction C cells as well, although with a reduced efficiency. However, eYFP+ fraction B cells from Sox4fl/fl;Rosa26eYFP mice (ie, with both copies of the Sox4 gene deleted) remained as fraction B cells and yielded no fraction C cells. These observations clearly demonstrated the essential role of Sox4 in controlling the differentiation of pro-B fraction B cells into fraction C cells.
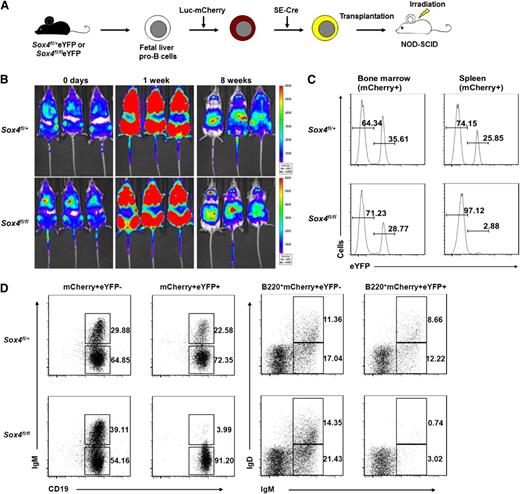
To test the validity of our new culture system, pro-B cells were labeled with luciferase-mCherry reporter and transduced with SE-Cre, and the resultant mixture of eYFP+ (with Sox4fl deletion) and eYFP− (without Sox4fl deletion) cells was transplanted in recipient mice. Immediately after transplantation, prompt homing of the transplanted cells to bone marrow was observed (0 days), and in 1 week posttransplantation, bone marrow luminescence signal intensity increased several fold. This finding indicated that the transplanted cells had undergone proliferation (Figure 3B). Notably, the major luminescent signal eventually shifted from bone marrow to the abdominal region in 8 weeks, an observation suggesting that maturation of the pro-B cells had occurred and they had migrated from the bone marrow to the spleen.
In vivo validation of the in vitro culture system reveals that loss of Sox4 impairs the development of pro-B cells into mature B cells. (A) Schematic representation of the transplantation model. Fetal liver pro-B cells were transduced with a dual reporter carrying luciferase (Luc), which permitted the tracing of cell development in vivo by bioimaging, and mCherry, which allowed analysis of transplanted cells by flow cytometry. Cells were transplanted into 250-rad-irradiated NOD-SCID mice 9 days after SE-Cre introduction. (B) Development of transplanted pro-B cells in NOD-SCID mice (Xenogen IVIS live imaging) at the indicated posttransplantation time points. Note the increase in the signal at the bone marrow position at 1 week and the shift of the signal to the spleen position at 8 weeks. Scale bars indicate signal intensity. (C) Analysis of transplanted pro-B cells from the bone marrow and spleen of the recipient mice. Bone marrow and spleen lymphocytes were analyzed by flow cytometry for eYFP expression 10 days posttransplantation. (D) Flow cytometry analysis of B cells from the bone marrow of recipient mice 10 days posttransplantation. mCherry+ B220+CD19+ cells were examined for expression of IgM and IgD. eYFP− cells (without Sox4 deletion) served as an internal control. Numbers indicate percent of cells (C-D). Data are representative of 4 independent experiments (B-D; n = 18 mice).
In vivo validation of the in vitro culture system reveals that loss of Sox4 impairs the development of pro-B cells into mature B cells. (A) Schematic representation of the transplantation model. Fetal liver pro-B cells were transduced with a dual reporter carrying luciferase (Luc), which permitted the tracing of cell development in vivo by bioimaging, and mCherry, which allowed analysis of transplanted cells by flow cytometry. Cells were transplanted into 250-rad-irradiated NOD-SCID mice 9 days after SE-Cre introduction. (B) Development of transplanted pro-B cells in NOD-SCID mice (Xenogen IVIS live imaging) at the indicated posttransplantation time points. Note the increase in the signal at the bone marrow position at 1 week and the shift of the signal to the spleen position at 8 weeks. Scale bars indicate signal intensity. (C) Analysis of transplanted pro-B cells from the bone marrow and spleen of the recipient mice. Bone marrow and spleen lymphocytes were analyzed by flow cytometry for eYFP expression 10 days posttransplantation. (D) Flow cytometry analysis of B cells from the bone marrow of recipient mice 10 days posttransplantation. mCherry+ B220+CD19+ cells were examined for expression of IgM and IgD. eYFP− cells (without Sox4 deletion) served as an internal control. Numbers indicate percent of cells (C-D). Data are representative of 4 independent experiments (B-D; n = 18 mice).
To examine the exact differentiation status of transplanted pro-B cells, we harvested bone marrow and spleen cells 10 days posttransplantation and analyzed them by flow cytometry. The number of Sox4fl/fleYFP+ cells was slightly lower in bone marrow but dramatically lower in the spleen than the corresponding numbers of Sox4fl/+eYFP+ cells. These differences suggested that Sox4 deletion affected the maturation of pro-B cells in the bone marrow and indirectly affected the number of cells that had egressed from bone marrow and migrated to the spleen (Figure 3C). We then stained the bone marrow cells for IgM and IgD, which are definitive markers for normal immature (IgM+IgD−) and mature (IgM+IgD+) B cells. Only a mild reduction (∼20%) was observed in Sox4fl/+eYFP+ cells, whereas a reduction of ∼90% was detected in CD19+IgM+ and in B220+IgM+IgD+Sox4fl/fleYFP+ cells compared with their respective eYFP− cells (Figure 3D). These results indicated that the cultured pro-B cells with functional Sox4 could develop into immature and mature B cells in vivo, but those with Sox4-depleted cells could not. The results also justify the potential utility of our system in studying the role of Sox4 and its downstream genes.
Identification and characterization of the Sox4 transcriptional network by both loss of function and gain of function
In our initial search, we had examined the possibility of a role for the known B-cell-specific genes in the Sox4 downstream network (supplemental Figure 1). Subsequently, to systematically identify a Sox4-controlled transcriptional program, we performed gene expression microarray analysis for both loss of function and gain of function of Sox4 by using cells from our newly established culture system. For loss of function, we compared the expression profiles of fraction B Sox4fl/flSE-Cre and Sox4fl/fl cells that were sorted out 9 days after the introduction of SE-Cre. For gain of function, we generated p190 Bcr-Abl-immortalized cells that had ectopic expression of hemagglutinin (HA)–tagged Sox4 (HA-Sox4) (the HA tag was used as a control) and a deletion of floxed Sox4 (Sox4fl/flSE-Cre) (Figure 4A-C). Importantly, in p190 Bcr-Abl-immortalized pro-B cells with Sox4 gene deletion, differentiation from fraction B to fraction C was restored with expression of HA-Sox4 (Figure 4D), suggesting that HA-Sox4 possessed normal Sox4 function in this process and the immortalized pro-B cells responded to the Sox4 function in the same manner as did normal primary pro-B cells.
Identification and characterization of the Sox4 transcriptional network. (A) Schematic representation of HA-Sox4 complementation experiments that required sequential transduction of 3 viral vectors. After each step, we sorted out cells positive for appropriate markers by flow cytometry. First, cultured fetal liver pro-B cells from Sox4fl/fl;Rosa26eYFP mouse embryos were immortalized by p190 Bcr-Abl (with mCherry as a marker). Second, SE-Cre was introduced, resulting in a deletion of floxed Sox4 and the STOP cassette before the eYFP sequence, and the cells became eYFP+. Finally, HA-Sox4 or HA (with mCerulean as a marker) was introduced. The reason we used p190 Bcr-Abl to immortalize the pro-B cells was that ectopic expression of Sox4 in primary pro-B cells instantaneously impeded cell survival, and immortalization of pro-B cells with p190 Bcr-Abl could overcome this problem. Moreover, this sequential introduction of vectors minimized potential confounding effects of BCR-ABL as the HA-Sox4-expressing experimental cells and HA-expressing control cells were generated from the same group of BCR-ABL+ cells. (B) Immunoblot analysis of Sox4 expression in HA-Sox4- or HA-transduced and immortalized pro-B cells that were depleted of endogenous Sox4. α-Tubulin served as a loading control. (C) Transcription activation by HA-Sox4 in luciferase reporter assays of 293T cells (error bars, standard error of the mean [SEM]). (D) Effect of HA-Sox4 expression on the differentiation from fraction B to fraction C of immortalized pro-B cells with a deletion of endogenous Sox4 (Sox4fl/flSE-Cre). Cells were analyzed by flow cytometry 7 days after introduction of HA or HA-Sox4. Numbers indicate percentages of cells in each quadrant. (E) Heat map of gene expression microarray profiling of cultured pro-B cells for loss of function (left, Sox4fl/fl vs Sox4fl/flSE-Cre) and gain of function (right, Sox4fl/flSE-Cre HA vs Sox4fl/flSE-Cre HA-Sox4). Expression of the top 100 differentially expressed genes (log2 fold change exceeding ±0.75 and P < .01) is depicted. Scale bar, intensity of signal. Each column represents an independent sample, and each row represents a gene. (F) Overlap between gene expression microarrays for loss (blue oval) and gain (yellow oval) of function of Sox4. Black and red numbers indicate down- and upregulated genes, respectively, in the loss-of-function array and vice versa in the gain-of-function array. (G-H) Ingenuity pathway analysis of gene expression profiles from the Sox4 gain-of-function array. Genes that showed a log2 fold change exceeding ±1.5 were analyzed. The functions of the differentially expressed genes in development (G) and canonical signaling pathways (H) are shown. Data are representative of 2 (B-C), 3 (D), or 1 (E-H) experiment.
Identification and characterization of the Sox4 transcriptional network. (A) Schematic representation of HA-Sox4 complementation experiments that required sequential transduction of 3 viral vectors. After each step, we sorted out cells positive for appropriate markers by flow cytometry. First, cultured fetal liver pro-B cells from Sox4fl/fl;Rosa26eYFP mouse embryos were immortalized by p190 Bcr-Abl (with mCherry as a marker). Second, SE-Cre was introduced, resulting in a deletion of floxed Sox4 and the STOP cassette before the eYFP sequence, and the cells became eYFP+. Finally, HA-Sox4 or HA (with mCerulean as a marker) was introduced. The reason we used p190 Bcr-Abl to immortalize the pro-B cells was that ectopic expression of Sox4 in primary pro-B cells instantaneously impeded cell survival, and immortalization of pro-B cells with p190 Bcr-Abl could overcome this problem. Moreover, this sequential introduction of vectors minimized potential confounding effects of BCR-ABL as the HA-Sox4-expressing experimental cells and HA-expressing control cells were generated from the same group of BCR-ABL+ cells. (B) Immunoblot analysis of Sox4 expression in HA-Sox4- or HA-transduced and immortalized pro-B cells that were depleted of endogenous Sox4. α-Tubulin served as a loading control. (C) Transcription activation by HA-Sox4 in luciferase reporter assays of 293T cells (error bars, standard error of the mean [SEM]). (D) Effect of HA-Sox4 expression on the differentiation from fraction B to fraction C of immortalized pro-B cells with a deletion of endogenous Sox4 (Sox4fl/flSE-Cre). Cells were analyzed by flow cytometry 7 days after introduction of HA or HA-Sox4. Numbers indicate percentages of cells in each quadrant. (E) Heat map of gene expression microarray profiling of cultured pro-B cells for loss of function (left, Sox4fl/fl vs Sox4fl/flSE-Cre) and gain of function (right, Sox4fl/flSE-Cre HA vs Sox4fl/flSE-Cre HA-Sox4). Expression of the top 100 differentially expressed genes (log2 fold change exceeding ±0.75 and P < .01) is depicted. Scale bar, intensity of signal. Each column represents an independent sample, and each row represents a gene. (F) Overlap between gene expression microarrays for loss (blue oval) and gain (yellow oval) of function of Sox4. Black and red numbers indicate down- and upregulated genes, respectively, in the loss-of-function array and vice versa in the gain-of-function array. (G-H) Ingenuity pathway analysis of gene expression profiles from the Sox4 gain-of-function array. Genes that showed a log2 fold change exceeding ±1.5 were analyzed. The functions of the differentially expressed genes in development (G) and canonical signaling pathways (H) are shown. Data are representative of 2 (B-C), 3 (D), or 1 (E-H) experiment.
Loss-of-function microarray profiling identified 247 downregulated genes and 126 upregulated genes in Sox4-deficient cells. This finding indicated that Sox4 functions predominantly as a transactivator rather than a transrepressor (Figure 4E, left). Gain-of-function microarray profiling identified 653 upregulated genes and 307 downregulated genes (Figure 4E, right). Importantly, 123 genes whose expression was downregulated upon Sox4 gene deletion were transcriptionally reinduced by expression of HA-Sox4. Likewise, 31 genes that were upregulated upon the loss of Sox4 underwent downregulation upon HA-Sox4 expression (Figure 4F). These results confirmed the involvement of these genes in the Sox4-controlled expression network. Ingenuity pathway analysis of the differentially expressed genes revealed their involvement in hematologic system development, particularly lymphoid system development (Figure 4G and supplemental Figure 2A). Interestingly, the Wnt/β-catenin pathway was identified as the one in which the Sox4 downstream genes were involved the most (Figure 4H and supplemental Figure 2B).
Comprehensive genome-wide identification and characterization of Sox4-binding elements
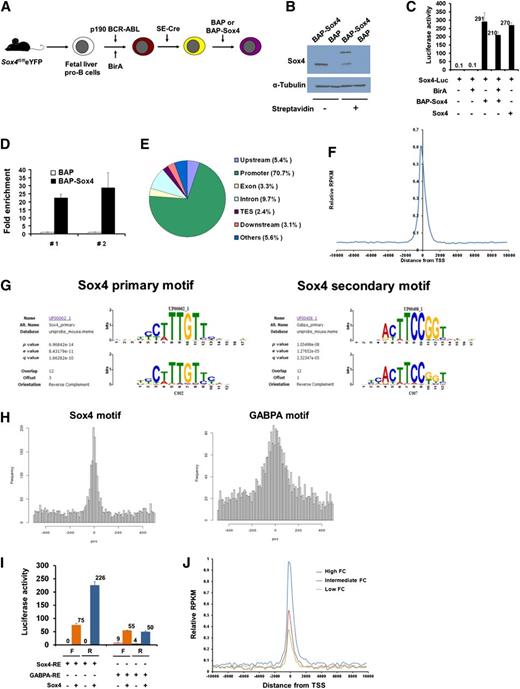
To systematically identify Sox4-binding elements, we adopted the newly developed in vitro biotinylation technique, bioChIP.35 We introduced the biotin-conjugating enzyme BirA ligase and biotin acceptor peptide (BAP)–tagged Sox4 (BAP-Sox4) (BAP served as a control) into the p190 Bcr-Abl-immortalized cells with Sox4fl/fl deleted (Figure 5A). We verified that biotinylated BAP-Sox4 could bind streptavidin, possessed Sox4 transcription activity, and was able to enrich Sox4-binding motifs (Figure 5B-D). These experiments demonstrated the validity of the bioChIP system in pulling down Sox4-binding elements.
Genome-wide identification and characterization of Sox4-binding elements. (A) Schematic outline of the experimental model, in which fetal liver pro-B cells were immortalized, transduced with BirA ligase, Sox4-depleted, and transduced with BAP-Sox4 or BAP. (B) Immunoblot analysis of Sox4 in BAP-Sox4- or BAP-transduced pro-B cells, which were immortalized, transduced with BirA, and depleted of endogenous Sox4. BAP-Sox4 had bound to streptavidin and yielded an additional slow-migrating band. α-Tubulin served as a loading control. (C) Activation of Sox4-luciferase reporter gene expression by biotinylated BAP-Sox4 (in the presence of BirA ligase) in 293T cells. (D) Enrichment of Sox4-binding elements in bioChIP. The multimerized Sox4 canonical motif was introduced into BAP or BAP-Sox4 pro-B cells and examined for changes in bioChIP DNA by quantitative PCR (qPCR). The amount of bioChIP DNA detected in BAP cells was set as 1. #1 and #2 denote results from 2 different sets of the primers. (E) Pie diagram of the genome-wide distribution of Sox4-binding regions in pro-B cells. (F) Enrichment of the Sox4 signal over the transcription start sites (TSS) of gene promoters. (G) Identification of Sox4-binding motifs by CisFinder. Sox4 canonical motif from the UniProbe database and the Sox4 motif identified in this study (left). GA-binding protein α chain (GABPA) canonical motif from the UniProbe database and the Sox4 motif identified in this study (right). (H) Histograms of the distances from the Sox4 (left) and GABPA (right) binding motifs to their peak summits. (I) Sox4 transactivation function on canonical Sox4 (8 copies) and GABPA (4 copies) motifs, cloned in either the forward or the reverse orientation, in luciferase reporter assays of 293T cells. (J) Association between Sox4-binding signals and the concomitant gene expression. A total of 16 984 genes present in the gain-of-function microarray and the RefSeq database were ranked by log2 fold change (FC) in ascending order, and genes with low (1-1000), intermediate (7992-8991), and high (15 985-16 984) levels of change were analyzed for Sox4-binding signals surrounding the 10-kb region of the transcriptional start site. Data are representative of 2 (B-C,I), 3 (D), or 1 (E-H,J) experiment; error bars (C-D,I), SEM. RPKM, reads per kilobase per million tags.
Genome-wide identification and characterization of Sox4-binding elements. (A) Schematic outline of the experimental model, in which fetal liver pro-B cells were immortalized, transduced with BirA ligase, Sox4-depleted, and transduced with BAP-Sox4 or BAP. (B) Immunoblot analysis of Sox4 in BAP-Sox4- or BAP-transduced pro-B cells, which were immortalized, transduced with BirA, and depleted of endogenous Sox4. BAP-Sox4 had bound to streptavidin and yielded an additional slow-migrating band. α-Tubulin served as a loading control. (C) Activation of Sox4-luciferase reporter gene expression by biotinylated BAP-Sox4 (in the presence of BirA ligase) in 293T cells. (D) Enrichment of Sox4-binding elements in bioChIP. The multimerized Sox4 canonical motif was introduced into BAP or BAP-Sox4 pro-B cells and examined for changes in bioChIP DNA by quantitative PCR (qPCR). The amount of bioChIP DNA detected in BAP cells was set as 1. #1 and #2 denote results from 2 different sets of the primers. (E) Pie diagram of the genome-wide distribution of Sox4-binding regions in pro-B cells. (F) Enrichment of the Sox4 signal over the transcription start sites (TSS) of gene promoters. (G) Identification of Sox4-binding motifs by CisFinder. Sox4 canonical motif from the UniProbe database and the Sox4 motif identified in this study (left). GA-binding protein α chain (GABPA) canonical motif from the UniProbe database and the Sox4 motif identified in this study (right). (H) Histograms of the distances from the Sox4 (left) and GABPA (right) binding motifs to their peak summits. (I) Sox4 transactivation function on canonical Sox4 (8 copies) and GABPA (4 copies) motifs, cloned in either the forward or the reverse orientation, in luciferase reporter assays of 293T cells. (J) Association between Sox4-binding signals and the concomitant gene expression. A total of 16 984 genes present in the gain-of-function microarray and the RefSeq database were ranked by log2 fold change (FC) in ascending order, and genes with low (1-1000), intermediate (7992-8991), and high (15 985-16 984) levels of change were analyzed for Sox4-binding signals surrounding the 10-kb region of the transcriptional start site. Data are representative of 2 (B-C,I), 3 (D), or 1 (E-H,J) experiment; error bars (C-D,I), SEM. RPKM, reads per kilobase per million tags.
As only a negligible amount of chromatin could be precipitated from BAP cells, input chromatin DNA from BAP-Sox4 cells was used as a control for next-generation sequencing. Our results showed that ∼70% of the sequence tags in both the input control and the ChIP DNA samples were able to map to unique locations in the mouse genome. Peaks were identified by using the Model-based Analysis of ChIP-Seq program and setting a P value of .00001 with a window size of 300 bp as a cutoff for calling significant peaks. In Sox4 bioChIP DNA, 8730 peaks were identified, but, with the same criteria, only 10 peaks were called in input control DNA. This result indicated that the peaks in Sox4 bioChIP DNA are highly specific. In the BAP-Sox4 ChIP DNA sample, most peaks were present in promoter regions (Figure 5E). Interestingly, 63.5% of the peaks coincided with transcription start sites (Figure 5F).
CisFinder analyses of sequences within 200 bp of the summit of all peaks identified 8 known transcription factor binding motifs.39 The identified motifs were further fine-tuned by the CisFinder resampling method (supplemental Figure 3A). We found the canonical motif of Sox4 (primary) to be the most significant motif identified by CisFinder. Among the 8730 total Sox4 peaks identified, 3007 contained this canonical motif (Motif Alignment & Search Tool, P ≤ .0001), and in 1575 peaks the canonical motif was found within 200 bp from the summit of the peaks (supplemental Table 1). Among the other 7 transcription factor motifs identified, a motif that shares a sequence with the GABPA motif was significantly enriched in the summits of the peaks (Figure 5G-H; supplemental Table 1 and supplemental Figure 3B). We further demonstrated that Sox4 was able to transactivate the GABPA motif in both orientations in reporter assays, although the efficiency was lower than that for the Sox4 primary motif (Figure 5I). These results suggested that Sox4 can orchestrate its transactivation function through the GABPA motif in early B-cell development. We also compared Sox4-binding signals among 3 groups of genes from the gain-of-function microarray with low, intermediate, and high levels of change in expression and found that the intensity of Sox4-binding signals was positively correlated with the levels of gene expression. This finding provided further evidence of direct involvement of Sox4 in transcriptional regulation of these genes (Figure 5J).
Downregulation of Rag1/2 gene expression and deficient Igh gene recombination in the absence of Sox4
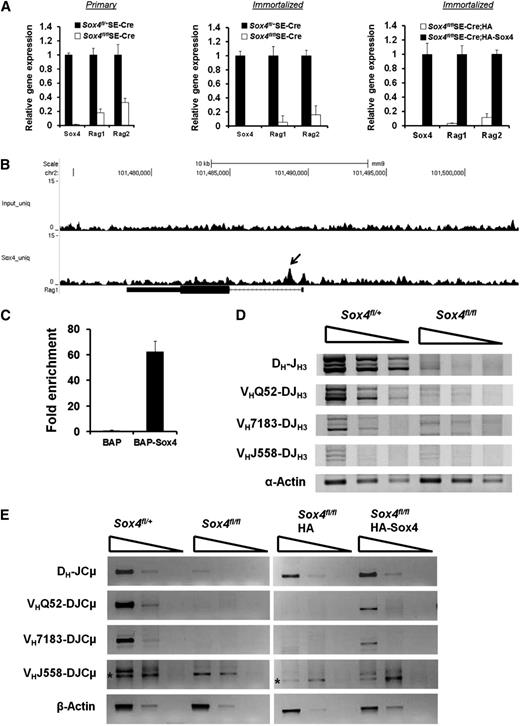
VDJ recombination at the Igh locus mediated by Rag1 and Rag2 is a prerequisite for the transition of pro-B cells to pre-B cells.12,13,16 Our gene expression microarray analysis identified Rag1 to be one of the genes mostly downregulated upon Sox4 depletion. We further demonstrated that Rag1 and Rag2 expression levels in primary or immortalized Sox4fl/flSE-Cre pro-B cells were significantly lower than those in Sox4fl/+SE-Cre pro-B cells. Moreover, HA-Sox4 expression in pro-B cells with Sox4fl/fl deleted upregulated the levels of Rag1 and Rag2 32- and 9-fold, respectively (Figure 6A). In addition, our ChIP-Seq analysis located a Sox4-binding element in the intron of Rag1, and qPCR analysis indicated 62-fold enrichment of this Sox4-binding sequence in Sox4-expressing cells as compared with control cells (Figure 6B-C). In agreement with the downregulation of Rag1 and Rag2 in Sox4-deficient pro-B cells, we detected reduced frequencies of recombination between DH-J, VHQ52-DJ, VH7183-DJ, and VH558-DJ gene segments (Figure 6D). The expression levels of recombined DH-JCμ, VHQ52-DJCμ, VH7183-DJCμ, and VH558-DJCμ gene products were also significantly reduced in comparison with the levels in Sox4fl/+SE-Cre pro-B cells. In contrast, expression of HA-Sox4 in pro-B cells with Sox4fl/fl deletion restored not only Rag1 and Rag2 expression but also the recombination events (Figure 6E). These data suggested that Sox4 controls the recombination of VHDJ genes in pro-B cells by regulating the expression of Rag1 and Rag2.
Sox4 induces the expression of Rag1 and Rag2 and controls gene recombination at the Igh loci. (A) Real-time RT-PCR analysis of Sox4, Rag1, and Rag2 expression in cDNAs prepared from primary (left) or immortalized (middle) Sox4fl/+SE-Cre and Sox4fl/flSE-Cre pro-B cells or from HA- or HA-Sox4-expressing and immortalized pro-B cells that had the endogenous floxed Sox4 deletion (right). Fraction B cells that were sorted out 9 days after the introduction of SE-Cre were used in the analysis (left and middle). In each pair, the higher level was set as 1 (error bars, SEM). (B) Genome browser view of Sox4 binding in the Rag1 locus identified by ChIP-Seq. Rag1 transcript in relation to Sox4 binding is shown. The arrow points to the Sox4-binding site. chr, chromosome. (C) qPCR detection of the Sox4 binding to Rag1 locus. Sox4 binding to the first intron of Rag1 was detected by qPCR amplification of bioChIP DNA from BAP or BAP-Sox4 cells, and the results were normalized to those from corresponding input DNA samples (normalized results from BAP cells were set as 1). (D) Semiquantitative PCR analysis of DH-JH3, VHQ52-DJH3, VH7183-DJH3, and VHJ558-DJH3 gene rearrangements in fivefold serial dilutions of DNA prepared from Sox4fl/+SE-Cre and Sox4fl/flSE-Cre pro-B cells. α-Actin served as a normalization control. (E) Semiquantitative RT-PCR analysis of rearranged DH-JCμ, VHQ52-DJCμ, VH7183-DJCμ, and VHJ558-DJCμ transcripts in fivefold serial dilutions of cDNA prepared from Sox4-deficient pro-B cells (left) and HA-Sox4-complemented pro-B cells (right). * denotes nonspecific amplification. β-Actin served as a normalization control. Data are representative of 1 (B-C) or 3 (A,D-E) independent experiments.
Sox4 induces the expression of Rag1 and Rag2 and controls gene recombination at the Igh loci. (A) Real-time RT-PCR analysis of Sox4, Rag1, and Rag2 expression in cDNAs prepared from primary (left) or immortalized (middle) Sox4fl/+SE-Cre and Sox4fl/flSE-Cre pro-B cells or from HA- or HA-Sox4-expressing and immortalized pro-B cells that had the endogenous floxed Sox4 deletion (right). Fraction B cells that were sorted out 9 days after the introduction of SE-Cre were used in the analysis (left and middle). In each pair, the higher level was set as 1 (error bars, SEM). (B) Genome browser view of Sox4 binding in the Rag1 locus identified by ChIP-Seq. Rag1 transcript in relation to Sox4 binding is shown. The arrow points to the Sox4-binding site. chr, chromosome. (C) qPCR detection of the Sox4 binding to Rag1 locus. Sox4 binding to the first intron of Rag1 was detected by qPCR amplification of bioChIP DNA from BAP or BAP-Sox4 cells, and the results were normalized to those from corresponding input DNA samples (normalized results from BAP cells were set as 1). (D) Semiquantitative PCR analysis of DH-JH3, VHQ52-DJH3, VH7183-DJH3, and VHJ558-DJH3 gene rearrangements in fivefold serial dilutions of DNA prepared from Sox4fl/+SE-Cre and Sox4fl/flSE-Cre pro-B cells. α-Actin served as a normalization control. (E) Semiquantitative RT-PCR analysis of rearranged DH-JCμ, VHQ52-DJCμ, VH7183-DJCμ, and VHJ558-DJCμ transcripts in fivefold serial dilutions of cDNA prepared from Sox4-deficient pro-B cells (left) and HA-Sox4-complemented pro-B cells (right). * denotes nonspecific amplification. β-Actin served as a normalization control. Data are representative of 1 (B-C) or 3 (A,D-E) independent experiments.
Downregulation of CK1ε gene expression and increased Wnt/β-catenin activity in the absence of Sox4
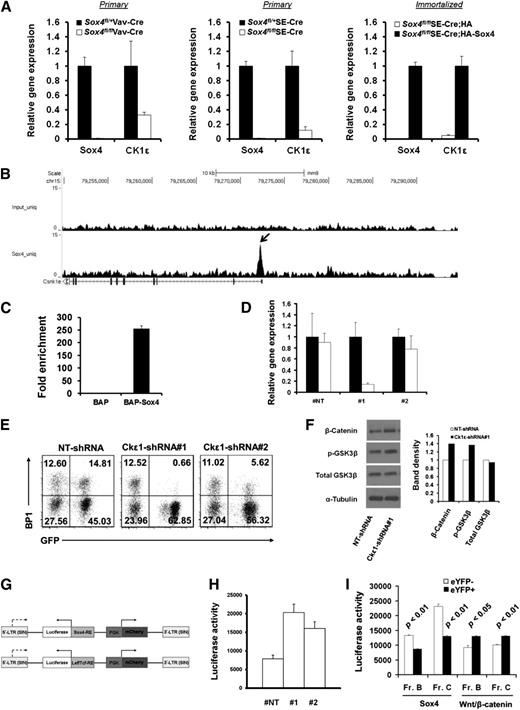
Ingenuity pathway analysis of gene expression microarray data suggested involvement of Sox4-regulated genes in multiple signaling pathways, with the Wnt/β-catenin signaling pathway having the highest level of involvement (Figure 4H and supplemental Figure 2B). In agreement with this analysis, our gene expression microarray studies identified CK1ε, which encodes a critical kinase in the Wnt/β-catenin pathway, to be one of the most differentially regulated genes.40,41 We next determined the level of CK1ε expression and that of Sox4 expression in both bone marrow and cultured pro-B cells. The level of CK1ε was lower in residual pro-B cells from Sox4fl/flVav-Cre mice and in Sox4fl/flSE-Cre pro-B cells than in the counterpart control cells. As expected, complementation with HA-Sox4 in pro-B cells with Sox4fl/fl deletion resulted in a 21-fold upregulation of CK1ε expression (Figure 7A). Our genome-wide search for Sox4-binding elements through ChIP-Seq analysis revealed binding of Sox4 to the proximal promoter of CK1ε, and qPCR analysis indicated 255-fold enrichment of this Sox4-binding sequence in Sox4-expressing cells as compared with control cells (Figure 7B-C). These results demonstrated that Sox4 is a critical positive regulator of CK1ε in pro-B cells.
Sox4 induces CK1ε expression and negatively regulates Wnt/β-catenin signaling. (A) Real-time RT-PCR analysis of Sox4 and CK1ε expression in pro-B cells sorted from the bone marrow of Sox4fl/+Vav-Cre and Sox4fl/flVav-Cre mice (left), fraction B Sox4fl/+ and Sox4fl/fl pro-B cells sorted from cultures of 9 days post-SE-Cre-transduction (middle), and HA- or HA-Sox4-expressing p190 Bcr-Abl-immortalized pro-B cells that had the endogenous floxed Sox4 deletion (right). In each pair, the higher expression was set as 1. (B) Genome browser view of Sox4 binding in the CK1ε locus identified by ChIP-Seq. CK1ε transcript in relation to Sox4 binding is shown. The arrow points to the identified Sox4-binding site. (C) qPCR detection of the Sox4 binding to CK1ε locus. Sox4 binding to proximal promoter of CK1ε locus was detected by qPCR amplification of bioChIP DNA from BAP or BAP-Sox4 cells, and the results were normalized to those from corresponding input DNA samples (normalized results from BAP cells were set as 1). (D) Real-time RT-PCR analysis of CK1ε expression in pro-B cells transduced with NT-shRNA or 2 different shRNAs specific for CK1ε. Expression in nontransduced cells was set as 1. (E) Effect of CK1ε silencing in pro-B cells on the transition from fraction B to fraction C. Fraction B cells were sorted out after 3 weeks of NT or CK1ε shRNA transduction and cultured for an additional 6 days before flow cytometry analysis. Numbers indicate percentages of cells in each quadrant. (F) Immunoblot analysis of the levels of β-catenin, p-GSK3β, and GSK3β in total cell lysates from NT or CK1ε shRNA-expressing pro-B cells. α-Tubulin served as a loading control. The histogram shows the relative band intensity normalized to that of α-tubulin. (G) Schematic representation of dual reporter vectors. PGK promoter drives the expression of mCherry in the forward orientation, and Sox4 or β-catenin interacting Tcf elements drive luciferase reporter expression in the opposite orientation. LTR, long terminal repeat; RE, response element; SIN, self-inactivating (see supplemental Methods for details of vector construction). (H) Effect of CK1ε silencing on the Wnt/β-catenin luciferase reporter activity in pro-B cells. (I) Sox4 and Wnt/β-catenin luciferase reporter activity in pro-B cells with or without Sox4 deletion. Data are representative of 1 (B-C), 2 (F), or 3 (A,D-E,H-I) independent experiments; error bars (A,C-D,H-I), SEM.
Sox4 induces CK1ε expression and negatively regulates Wnt/β-catenin signaling. (A) Real-time RT-PCR analysis of Sox4 and CK1ε expression in pro-B cells sorted from the bone marrow of Sox4fl/+Vav-Cre and Sox4fl/flVav-Cre mice (left), fraction B Sox4fl/+ and Sox4fl/fl pro-B cells sorted from cultures of 9 days post-SE-Cre-transduction (middle), and HA- or HA-Sox4-expressing p190 Bcr-Abl-immortalized pro-B cells that had the endogenous floxed Sox4 deletion (right). In each pair, the higher expression was set as 1. (B) Genome browser view of Sox4 binding in the CK1ε locus identified by ChIP-Seq. CK1ε transcript in relation to Sox4 binding is shown. The arrow points to the identified Sox4-binding site. (C) qPCR detection of the Sox4 binding to CK1ε locus. Sox4 binding to proximal promoter of CK1ε locus was detected by qPCR amplification of bioChIP DNA from BAP or BAP-Sox4 cells, and the results were normalized to those from corresponding input DNA samples (normalized results from BAP cells were set as 1). (D) Real-time RT-PCR analysis of CK1ε expression in pro-B cells transduced with NT-shRNA or 2 different shRNAs specific for CK1ε. Expression in nontransduced cells was set as 1. (E) Effect of CK1ε silencing in pro-B cells on the transition from fraction B to fraction C. Fraction B cells were sorted out after 3 weeks of NT or CK1ε shRNA transduction and cultured for an additional 6 days before flow cytometry analysis. Numbers indicate percentages of cells in each quadrant. (F) Immunoblot analysis of the levels of β-catenin, p-GSK3β, and GSK3β in total cell lysates from NT or CK1ε shRNA-expressing pro-B cells. α-Tubulin served as a loading control. The histogram shows the relative band intensity normalized to that of α-tubulin. (G) Schematic representation of dual reporter vectors. PGK promoter drives the expression of mCherry in the forward orientation, and Sox4 or β-catenin interacting Tcf elements drive luciferase reporter expression in the opposite orientation. LTR, long terminal repeat; RE, response element; SIN, self-inactivating (see supplemental Methods for details of vector construction). (H) Effect of CK1ε silencing on the Wnt/β-catenin luciferase reporter activity in pro-B cells. (I) Sox4 and Wnt/β-catenin luciferase reporter activity in pro-B cells with or without Sox4 deletion. Data are representative of 1 (B-C), 2 (F), or 3 (A,D-E,H-I) independent experiments; error bars (A,C-D,H-I), SEM.
We tested the role of CK1ε in B-cell development with small hairpin RNA (shRNA)–mediated knockdown. Real-time RT-PCR analysis showed 86% and 22% lower CK1ε expression levels in the cells containing #1 and #2 CK1ε-specific shRNA, respectively, and a 10% lower CK1ε expression level in the cells with nontargeting (NT) shRNA compared with the control (no shRNA) (Figure 7D). Notably, fraction B cells failed to differentiate into fraction C cells in vitro after knockdown of CK1ε expression (Figure 7E). These results indicated that the CK1ε deficiency had a blocking effect similar to that of the Sox4 deficiency on pro-B-cell differentiation, further suggesting that CK1ε is one of the Sox4-regulated genes.
CK1ε was reported to function as either a positive or a negative regulator of the Wnt/β-catenin pathway depending on cellular context.42 We therefore measured β-catenin levels and Wnt/β-catenin signaling activity in the cells with CK1ε knockdown. Elevated expression of β-catenin, as well as p-GSK3β, was detected by western blotting in the cells with CK1ε knockdown (Figure 7F). In agreement with this finding, we detected a 2.6-fold upregulation in Wnt/β-catenin reporter activity in CK1ε knockdown cells in comparison with the activity in NT-shRNA cells (Figure 7G-H). As expected, our reporter assay also demonstrated that decreased Sox4 activity in Sox4fl/flSE-Cre eYFP+ cells was accompanied by increased Wnt/β-catenin activity, in comparison with the activities in control eYFP− cells (Figure 7I). These findings suggest that CK1ε functions as a negative regulator of Wnt/β-catenin signaling in pro-B cells and Sox4 mediates downregulation of Wnt/β-catenin signaling by upregulating CK1ε.
Discussion
Our study has uncovered molecular mechanisms by which the transcription factor Sox4 regulates early B-cell development. Sox4 activates recombination of VHDJH genes by inducing the expression of Rag1 and Rag2 and mediates downregulation of Wnt/β-catenin signaling by inducing the expression of CK1ε, thus influencing these 2 indispensable processes in early B-cell development.
Sox4 is vital in early B-cell development.3,43 Sox4−/− embryonic liver cells fail to proliferate in the presence of IL-7 in vitro and to develop into mature B cells in vivo in transplantation experiments.2 The fact that Sox4-deficient B cells could not be obtained from culture or transplantation hindered further studies on the molecular function of Sox4 in B-cell development for many years. Likewise, in the work reported here, we failed to establish pro-B-cell cultures with fetal liver cells or bone marrow cells from Sox4fl/flVav-Cre mice, although we succeeded in culturing Sox4fl/+Vav-Cre pro-B cells. Therefore, it was necessary to develop a new system to obtain a sufficient number of Sox4-deficient pro-B cells for systematic molecular studies.
Our approach was to establish pro-B-cell cultures and then delete the Sox4 gene in vitro with Cre recombinase. Continuous expression of Cre recombinase appears to have overt toxicity and hampers cell survival (unpublished data).44 By using SE-Cre, which limits its own expression, we showed that Sox4 deletion was effectively induced without any toxicity to pro-B cells. This system allowed us to study, as Sox4 levels gradually decreased, the emerging phenotype of pro-B cells. Notably, sorted fraction B cells with Sox4 deficiency completely failed to differentiate into fraction C cells. We also showed that Sox4-deficient pre-pro-B cells failed to differentiate into pro-B cells in vitro, and pro-B cells were unable to develop into mature B cells after transplantation into irradiated recipient mice. Thus, Sox4 is involved in multiple stages of early B-cell development, from fraction A to fraction B, from fraction B to fraction C, and from fraction C to mature B cells.
The cultured Sox4fl/fl and Sox4fl/+ pro-B cells were not only unlimited in number, but also nearly 100% pure and only differed in the Sox4 expression and its associated traits, allowing us to systematically study the molecular alterations caused by loss of Sox4 and obtain unequivocal data. Early B-cell development is precisely controlled by a group of hierarchically ordered regulatory molecules, particularly transcription factors.5,45,46 For example, Pu.1, Ebf1, E2A, and Pax5 are involved in B-cell lineage determination and commitment. However, in Sox4-deficient pro-B cells, we observed normal expression levels of these genes, suggesting that Sox4 might control B-cell development by regulating a distinct gene network. In this study, we identified such a network in pro-B cells. Importantly, Ingenuity pathway analysis of Sox4-controlled genes revealed their involvement in the development of the lymphoid system. Our genome-wide search for Sox4-binding elements revealed that the elements exist predominantly within the proximal promoters of the genes. Furthermore, our motif search in the Sox4-binding regions identified the canonical Sox4 motif as the primary motif used by Sox4. We also found an additional Sox4-binding motif, which is known to be the native motif for the GABPA transcription factor. We observed transcriptional activation by Sox4 via the GABPA motif. Notably, loss of GABPA, like loss of Sox4, induces embryonic lethality and deficiency in pro-B cells.45,47
Rag1 and Rag2 are 2 critical components of VDJ recombinase, which mediates VDJ recombination at the Igh locus.12,13 In Rag1- or Rag2-deficient mice, B-cell development is blocked at the pro-B-cell stage.14-16 We noted that Rag1 and Rag2 expression levels were reduced and the rearrangements of Igh VDJ gene segments were markedly diminished in Sox4-deficient pro-B cells. In contrast, Rag1 and Rag2 expression and Igh VDJ rearrangements were restored in endogenous Sox4-depleted pro-B cells when the cells were complemented with ectopic Sox4 expression. Thus, Sox4 might control early B-cell development, at least in part, by regulating Rag1 and Rag2 expression and hence VDJ gene rearrangements at the Igh locus.
Ingenuity pathway analysis of Sox4-controlled genes identified the Wnt/β-catenin pathway as the pathway most influenced by Sox4. In agreement with this finding, we noted an increase in Wnt/β-catenin signaling activity in Sox4-deficient pro-B cells, suggesting that Sox4 might be one of the key negative regulators of Wnt/β-catenin signaling for normal B-cell development. Wnt/β-catenin signaling is known to have a critical function in HSCs. Expression of a constitutively activated form of β-catenin in HSCs compromised the ability of the HSCs to differentiate into multiple lineages, a finding suggesting that excessive Wnt/β-catenin signaling prevents HSCs from differentiating normally.28,29 On the other hand, conditional inactivation of β-catenin in B lymphoid progenitors has no effect on B-cell development.30,31 By loss- and gain-of-function microarray profiling, we identified CK1ε as one of the downstream mediators of Sox4. CK1ε, depending on cell context, has been reported to function as either a repressor or an activator of Wnt/β-catenin signaling.40-42 With CK1ε knockdown in pro-B cells, we observed upregulation of Wnt/β-catenin signaling and the transitional block from fraction B to fraction C, as seen with Sox4 deficiency. Thus, our findings suggest that Sox4 downregulates Wnt/β-catenin signaling by inducing CK1ε expression and such mechanistic regulation is important for induction of B lineage differentiation.
Our study has unraveled the molecular mechanisms underlying the role of Sox4 in early B-cell development. The fact that Sox4 is required at multiple stages of B-cell development reflects the importance of this protein. Sox4 exerts its function by suppressing differentiation-antagonizing Wnt/β-catenin signaling and activating the differentiation inducers Rag1 and Rag2. Further characterization of interaction between Sox4 and other transcription factors such as GABPA should shed more light on the molecular regulation of early B-cell development. The methods delineated in this study can be a resource for characterizing genes, like Sox4, whose deficiency is detrimental to early B-cell development.
There is an Inside Blood Commentary on this article in this issue.
The microarray and chromatin immunoprecipitation sequencing data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE50067).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank V. Lefebvre (Cleveland Clinic) for floxed Sox4 mice; D. Kioussis (Medical Research Council National Institute for Medical Research) for Vav-Cre mice; K. Dorshkind (University of California - Los Angeles Jonsson Comprehensive Cancer Center) for help with pro-B-cell culturing; S. M. Kim for help with microarray experiments; D. M. Livingston (Dana-Farber Cancer Institute) for the SE-Cre vector; H. Clever for the Sox4 reporter vector; P. Zweidler-McKay for the p190 Bcr-Abl expression vector; R. Nurieva for the mCerulean vector; K. Ramirez, K. Acklin, V. Papanna, K. Mandal, and K. Dwyer for help with flow cytometry sorting; L. Ramagli and K. Khanna for next-generation sequencing; H. Pelicano and X. Leng for discussions; A. Paranjape for help with figure preparation; S. Mani for critical review of the manuscript; and The University of Texas MD Anderson Cancer Center Department of Scientific Publications for editing the manuscript.
This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health grant 5R03AI079779-02 (X.S.), American Cancer Society Research Scholar Grant (X.S.), MD Anderson Cancer Center Physician Scientist Program (X.S.), MD Anderson Cancer Center Institutional Research Grant (X.S.), and MD Anderson Cancer Center Support Grant (CA016672).
Authorship
Contribution: S.M. and X.S. planned the study, designed the experiments, analyzed the data, and wrote the manuscript; S.M. did most of the experiments; B.S. generated Sox4fl/flVav-Cre mice and contributed to some other experiments; Y.L. performed ChIP-Seq analysis and contributed to the writing; H.M. contributed to VDJ recombination analysis; Y.G. and D.W. participated in designing and instructing the study; J.-S.L. and K.L. contributed to gene expression profiling experiments; and X.S. provided overall supervision of the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xiaoping Sun, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Unit 37, Houston, TX 77030; e-mail: xsun@mdanderson.org.

![Figure 4. Identification and characterization of the Sox4 transcriptional network. (A) Schematic representation of HA-Sox4 complementation experiments that required sequential transduction of 3 viral vectors. After each step, we sorted out cells positive for appropriate markers by flow cytometry. First, cultured fetal liver pro-B cells from Sox4fl/fl;Rosa26eYFP mouse embryos were immortalized by p190 Bcr-Abl (with mCherry as a marker). Second, SE-Cre was introduced, resulting in a deletion of floxed Sox4 and the STOP cassette before the eYFP sequence, and the cells became eYFP+. Finally, HA-Sox4 or HA (with mCerulean as a marker) was introduced. The reason we used p190 Bcr-Abl to immortalize the pro-B cells was that ectopic expression of Sox4 in primary pro-B cells instantaneously impeded cell survival, and immortalization of pro-B cells with p190 Bcr-Abl could overcome this problem. Moreover, this sequential introduction of vectors minimized potential confounding effects of BCR-ABL as the HA-Sox4-expressing experimental cells and HA-expressing control cells were generated from the same group of BCR-ABL+ cells. (B) Immunoblot analysis of Sox4 expression in HA-Sox4- or HA-transduced and immortalized pro-B cells that were depleted of endogenous Sox4. α-Tubulin served as a loading control. (C) Transcription activation by HA-Sox4 in luciferase reporter assays of 293T cells (error bars, standard error of the mean [SEM]). (D) Effect of HA-Sox4 expression on the differentiation from fraction B to fraction C of immortalized pro-B cells with a deletion of endogenous Sox4 (Sox4fl/flSE-Cre). Cells were analyzed by flow cytometry 7 days after introduction of HA or HA-Sox4. Numbers indicate percentages of cells in each quadrant. (E) Heat map of gene expression microarray profiling of cultured pro-B cells for loss of function (left, Sox4fl/fl vs Sox4fl/flSE-Cre) and gain of function (right, Sox4fl/flSE-Cre HA vs Sox4fl/flSE-Cre HA-Sox4). Expression of the top 100 differentially expressed genes (log2 fold change exceeding ±0.75 and P < .01) is depicted. Scale bar, intensity of signal. Each column represents an independent sample, and each row represents a gene. (F) Overlap between gene expression microarrays for loss (blue oval) and gain (yellow oval) of function of Sox4. Black and red numbers indicate down- and upregulated genes, respectively, in the loss-of-function array and vice versa in the gain-of-function array. (G-H) Ingenuity pathway analysis of gene expression profiles from the Sox4 gain-of-function array. Genes that showed a log2 fold change exceeding ±1.5 were analyzed. The functions of the differentially expressed genes in development (G) and canonical signaling pathways (H) are shown. Data are representative of 2 (B-C), 3 (D), or 1 (E-H) experiment.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/26/10.1182_blood-2013-12-543801/4/m_4064f4.jpeg?Expires=1769232928&Signature=yAVTYoLWIg2nN3RCQyxAzp4RSjCkOPf5dIu4LD700hGKRkKRQJOu6j9wBPemv6xAljvLzU86deobi5ocSMQdSS8uBITFmH0LMOqXTCIv7x9~3hwGeXJqSunpAxAvdgjlZq3czduw~56rKiCZDGoltjD8cWq0WaJrzXEwCNqAYV~4Q6RBT1Sth~gFXa8nXVT~mRP1UUq22IKHInWFovSWl~y~3jWKPR5-5SjE3Ryi7YOyb8eQQ030d89ez0zt5Y0VSftKBmrzP~h5AaCAxlOLFPEx6ewkfIRZLrK3duNpr9hP62rdkbh3umVXvSqBLczF38aPRy1fG8dswxPZM-f4jQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)