Key Points
Notch signals expand human HSC (CD90low) cells in vitro and delay the expansion of CD45RAint and CD45RAhi cells in vitro.
HSCs expanded in vitro are equal to ex vivo CD90low cells in immune reconstitution.
Abstract
All blood cell lineages start from hematopoietic stem cells (HSCs), which were recently shown to represent a heterogeneous group of cells. In mice, Notch signaling promotes the maintenance of “stemness” as well as the expansion of self-renewing HSCs in vitro. Additionally, human CD34+ cells were shown to expand in vitro in response to Notch signals. However, it is unclear whether Notch directly affects all HSCs, and whether this role is relevant in vivo. Here, we developed culture conditions that support the maintenance of CD34+CD133+CD90lowCD38−CD7−CD10−CD45RA− (CD90low) cells, phenotypically defined HSCs, as well as 2 early progenitor cells (CD34+CD38−CD7−CD10−CD45RAint [RAint] and CD34+CD38−CD7−CD10−CD45RAhi [RAhi]) that were functionally equivalent to multipotent progenitor-2 and lymphoid-primed multipotent progenitor, respectively, found in cord blood. Using a genetic approach, we show that Notch signals were required for HSC preservation, with cultured HSCs being equal to ex vivo HSC cells in their ability to reconstitute immunodeficient mice; however, dnMaml-transduced HSCs were not maintained in vitro. Interestingly, Notch signaling did not appear to be required for the self-renewal of human HSCs in vivo. Our findings support the notion that Notch signals maintain human HSCs in vitro that have hematopoietic-reconstituting ability in vivo and delay the appearance of 2 newly described early progenitor cells.
Introduction
The hematopoietic system has been conceptualized as operating within a hierarchical model, with the hematopoietic stem cell (HSC) at the apex.1 Human umbilical cord blood (CB) CD34− cells would appear to be the most quiescent HSCs capable of giving rise to CD34+CD90lowCD49f+ HSCs with long-term reconstituting ability.2-4 Whether HSCs are affected by Notch signaling remains controversial. For instance, Notch signaling does not appear to be required for the maintenance of adult HSCs in mice.5 In contrast, Notch signaling was shown to play an important role in the expansion of self-renewing HSCs in vitro.6
In humans, although a prominent role for Notch signaling is well documented based on the expansion of CD34+ cells in vitro, which includes both HSCs and progenitor cells, no quantitative and functional direct comparison in the efficiency of reconstitution to fresh ex vivo HSCs has been reported.7-13 Nonetheless, Notch appears to affect human HSCs because CD34−/+ HSCs were recently shown to have active Notch signaling. However, in contrast to the experimental approach used in mice, which shows that Notch is not required for HSC maintenance, no similar genetic manipulations have been reported for human HSCs.4,5
Several human progenitors able to differentiate into lymphomyeloid cells have been found in CB and/or bone marrow (BM).14-18 Nevertheless, it remains difficult to establish a clear hierarchical structure starting from an HSC. We sought to address this by starting cultures with purified CB-derived HSCs and functionally defined intermediates to yield a composite and hierarchical relationship. We modified the OP9-DL1 cell coculture system19,20 to support the expansion of HSCs and made use of OP9 cells expressing 2 different levels of Delta-like-4 (Dll4),21 therefore enabling the titration of Notch signaling on HSCs. Additionally, using a genetic approach, we found that Notch signaling during the coculture period was required for the maintenance of HSCs while delaying the appearance and frequency of downstream progenitors. However, in vivo competition experiments revealed that Notch signaling was not required for the maintenance of human HSCs within the BM.
Together, our findings support the notion that Notch signals can extend the period of time in which HSC function can be maintained in vitro by delaying the emergence of at least 2 early progenitors, which we have characterized here providing a more complete view of the early stages of human hematopoiesis.
Methods
Umbilical CB samples
Human CB samples were obtained by syringe extraction and collected in a blood-pack unit containing citrate phosphate dextrose anticoagulant (Baxter Healthcare) from consenting mothers following delivery in accordance with approved guidelines established by the research ethics board of Sunnybrook Health Sciences Centre. Within 24 hours of collection, CB mononuclear cells were isolated as previously described.19
Mice
NOD.cg-PrkdcscidIL2rgtm/Wjl/Sz (NOD/SCID/γcnull, NSG) and Rag2−/− γcnull BALB/c mice were purchased from The Jackson Laboratory and housed and bred in a pathogen-free facility. All animal procedures were approved by the Sunnybrook Health Science Centre Animal Care Committee.
Generation of OP9-Dll4
OP9-Ctrl, OP9-DL4hi, and OP9-DL4low cells were generated and maintained as previously described.22
CB purification
CB cells were lineage depleted using the Stem-Sep human progenitor cell enrichment kit (StemCell Technologies) according to the manufacturer’s protocol, and further purified as described in detail in the supplemental Methods (available on the Blood Web site).
Cocultures
OP9-Ctrl, OP9-DL4hi, and OP9-DL4low cells were plated and maintained as previously described,22-24 and used in cocultures as described in detail in the supplemental Methods.
For clonal analysis, sort-deposited cells were cultured for an additional 14 to 21 days under T or myeloid conditions as described in detail in the supplemental Methods.
In vivo reconstitutions
Donor cells were sorted for the indicated markers and then injected intrahepatically into neonatal immune-deficient mice irradiated with 2 Gy. After 10 weeks, mice were sacrificed and thymus, BM, and spleen were analyzed by flow cytometry. Details of the cells used and analysis are described in the supplemental Methods.
dnMaml construct and retroviral transductions
The sequence encoding for amino acids 8 to 74 from the mouse Mastermind like 1 (Maml1) gene was fused, in frame, to a 5′–enhanced green fluorescent protein (eGFP) sequence using described cloning methods. This dominant-negative form of MAML1 (dnMaml) was then cloned into retroviral or lentiviral vectors to be used for transduction experiments of CB cells as described in detail in the supplemental Methods.
Precursor-product relationships
In vitro–derived CD45RAint cells were sorted on day 18 from cultures, on OP9-Dl4hi-DL4low or -Ctrl. Sorted cells (10 000 cells) were then cultured again on the OP9 cells they were derived from, in Stem. After 2 and 3 days, analysis of CD45RA expression on gated CD34+CD38−CD7−CD10− cells was performed.
Quantitative polymerase chain reaction analysis
Total RNA was isolated in TRIzol reagent and reverse transcribed using Superscript-III and Oligo(dT) primers (Invitrogen). Diluted complementary DNA samples from cultures were used as described in detail in the supplemental Methods.
Statistical analysis
Student t unpaired and paired 2-tailed tests were performed using Excel and Prism software.
Results
Characterization of early hematopoietic subsets in CB
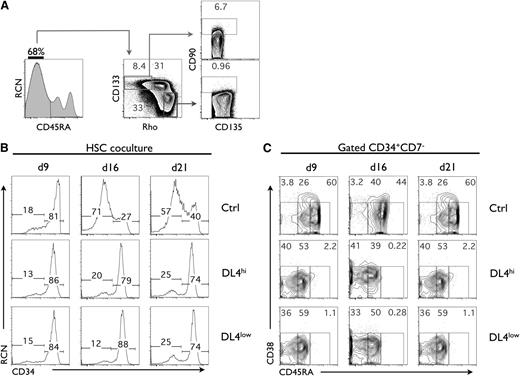
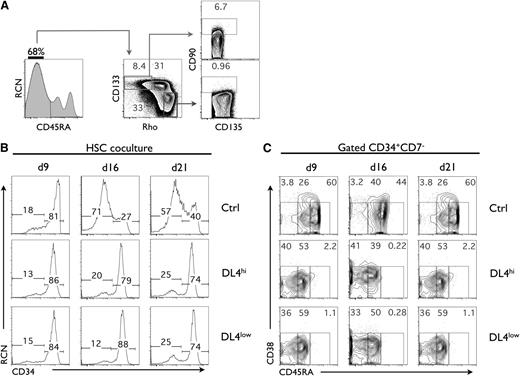
We have used CB to identify early hematopoietic subsets using previously described markers,2,25 with the addition of Rhodamine-123 (Rho) and CD133 to further characterize human HSC and multipotent progenitor (MPP) subsets. Analysis of CD45RA expression from Lin−CD135+ cells showed the presence of 3 populations, with the most abundant fraction being CD45RA−. As shown in Figure 1A, the CD45RA− fraction (CD45RA− cells) was further analyzed for CD133 expression and Rho uptake, and we identified the HSC fraction as having a CD90low phenotype, which was found only in the CD45RA−CD133+CD135+Rholow fraction. Not surprisingly, CD90− cells made up the vast majority of the Rholow subset, likely corresponding to an MPP-enriched subset. CD135+CD45RA−CD133+RholowCD90low cells, which were also CD38lowCD7−CD10− (not shown), hereafter referred to as HSCs, were used in subsequent experiments.
Notch signaling maintains a CD45RA−cell subset in cultures of umbilical CB HSCs. (A) Flow cytometric analysis of lineage-depleted CD135+. Three populations of CD45RA were observed (CD45RA−, CD45RAint, and CD45RAhi). CD45RA− were analyzed for CD133 expression and Rhodamine-1,2,3 (Rho) uptake which identified 3 levels of Rho staining (Rholow, Rhoint, Rhohi). The CD133+Rholow cells were analyzed for the expression of CD90 and CD135. CD90low are referred to as HSC, and CD90− cells are referred to as MPP. A representative experiment is shown from at least 10 independent analyses. (B) Cocultures were initiated with CD90low cells (HSCs) placed on OP9-Ctrl, -DL4hi, and -DL4low cells and analyzed by flow cytometry at days 9, 16, and 21 of culture for the expression of CD34 as indicated. (C) CD34+CD7− gated cells from the indicated cocultures were analyzed at days 9, 16, and 21 for the expression of CD38 and CD45RA. Numbers within each plot indicate the percentage of cells in the indicated gates. A representative experiment is shown, from at least 10 independent cultures.
Notch signaling maintains a CD45RA−cell subset in cultures of umbilical CB HSCs. (A) Flow cytometric analysis of lineage-depleted CD135+. Three populations of CD45RA were observed (CD45RA−, CD45RAint, and CD45RAhi). CD45RA− were analyzed for CD133 expression and Rhodamine-1,2,3 (Rho) uptake which identified 3 levels of Rho staining (Rholow, Rhoint, Rhohi). The CD133+Rholow cells were analyzed for the expression of CD90 and CD135. CD90low are referred to as HSC, and CD90− cells are referred to as MPP. A representative experiment is shown from at least 10 independent analyses. (B) Cocultures were initiated with CD90low cells (HSCs) placed on OP9-Ctrl, -DL4hi, and -DL4low cells and analyzed by flow cytometry at days 9, 16, and 21 of culture for the expression of CD34 as indicated. (C) CD34+CD7− gated cells from the indicated cocultures were analyzed at days 9, 16, and 21 for the expression of CD38 and CD45RA. Numbers within each plot indicate the percentage of cells in the indicated gates. A representative experiment is shown, from at least 10 independent cultures.
Cocultures with a modified OP9 system for HSC expansion
To examine how Notch signals could influence the CB HSC fraction, these cells were cultured on OP9-Ctrl cells (green fluorescent protein [GFP] alone) or on OP9 cells expressing different levels of Dll4 (OP9-DL4low and OP9-DL4hi)21 (supplemental Figure 1C), together with cytokines3,26 to support survival and expansion of HSCs and early progenitors. Cocultures were analyzed on a weekly basis for the expression of markers that define HSCs. By day 21, we noted that cocultures grown on OP9-DL4hi and -DL4low cells displayed a greater frequency of CD34+ cells than those on OP9-Ctrl cells (Figure 1B). An additional and expected effect of Notch signaling was the generation of CD34+CD7+ cells, early T-lineage cells,19 present in DL4hi and DL4low cocultures starting with either HSCs or MPPs, but not seen in the absence of Dll4 (control [Ctrl]) (supplemental Figure 1D).
Based on these observations, we looked specifically at CD34+CD7− cells at different time points, and found that CD45RA and CD38 were expressed at different levels and in different proportions on CD34+CD7− gated cells (Figure 1C). In particular, the CD45RA− and CD45RAint subsets were clearly present in cultures grown on DL4hi but not in Ctrl, while the CD45RAhi subset was abundantly present in cocultures lacking Notch signaling (Ctrl).
Because cocultures initiated with HSCs under Notch signaling conditions contained a substantial fraction of CD45RA− cells, we analyzed whether CD90low cells were present after 3 weeks. Supplemental Figure 1E shows CD90low cells only in DL4hi and DL4low cultures but not in OP9-Ctrl cultures, nor in cultures initiated with downstream CD90− cells (supplemental Figure 1F).
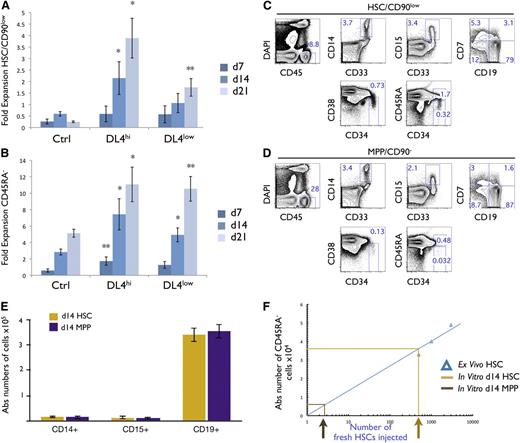
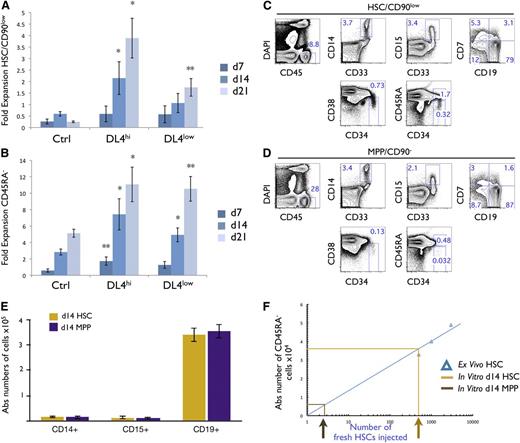
We calculated the expansion of CD45RA− (CD133+CD34+CD38−CD7−CD45RA−) cells, and phenotypically defined HSCs (pHSCs) as (CD133+CD34+CD38−CD7−CD45RA−CD90low) based on the absolute cellularity recovered from the starting HSCs (Figure 2A). CD45RA− and pHSCs cultured on Ctrl expanded minimally (Figure 2A-B). However, when cultured with DL4hi, CD45RA−, and pHSCs expanded sixfold to 12-fold and twofold to fourfold after 2 to 3 weeks, respectively (Figure 2A-B). A smaller, but significant, expansion was also seen with DL4low. Similar data were obtained when OP9-DL1hi cells were used (data not shown). These results suggest that a high or low level of Notch signaling retards differentiation of pHSCs, allowing for some cellular expansion. We conclude that the modified OP9-DL4 coculture system appears to maintain cells that are phenotypically identical to the input HSCs.
Notch signals lead to an enhanced cellular expansion of HSCs and CD45RA−cells with reconstitution potential in vivo. Cocultures were initiated with CD90low cells (HSCs) placed on OP9-Ctrl, -DL4hi, and -DL4low cells and analyzed by flow cytometry at days 7, 14, and 21 of culture. The cellular fold expansion from the input number (5-10 × 103) is shown for (A) CD90low and (B) CD45RA− cells. Bar graphs show mean and SE of 6 to 8 independent experiments. Statistical significance is indicated as (*) for P ≤ .05, and (**) for P ≤ .01. Cocultures were initiated with HSCs placed on OP9-DL4hi cells for 14 days, (C) phenotypically defined pHSC/CD90low, and (D) phenotypically defined pMPP/CD90− cells were isolated by flow cytometry, and 2 × 103 pHSC/CD90low and 5 × 104 pMPP/CD90− cells were injected into sublethally irradiated NSG neonatal mice. Ten weeks after injection, BM was analyzed by flow cytometry for the expression of the indicated marker. Numbers within each plot indicate the percentage of cells in the indicated gates or quadrants. (E) Absolute numbers of CD14+, CD15+, and CD19+ cells recovered. (F) A linear correlation between the absolute numbers of CD45RA− cells recovered from the BM and the number of freshly sorted HSCs injected into mice is shown (R2 = 0.87). Absolute numbers of CD45RA− cells recovered from (C) and (D) were plotted on the linear graph and the corresponding number of HSCs injected was extrapolated. A total of 8 mice were analyzed.
Notch signals lead to an enhanced cellular expansion of HSCs and CD45RA−cells with reconstitution potential in vivo. Cocultures were initiated with CD90low cells (HSCs) placed on OP9-Ctrl, -DL4hi, and -DL4low cells and analyzed by flow cytometry at days 7, 14, and 21 of culture. The cellular fold expansion from the input number (5-10 × 103) is shown for (A) CD90low and (B) CD45RA− cells. Bar graphs show mean and SE of 6 to 8 independent experiments. Statistical significance is indicated as (*) for P ≤ .05, and (**) for P ≤ .01. Cocultures were initiated with HSCs placed on OP9-DL4hi cells for 14 days, (C) phenotypically defined pHSC/CD90low, and (D) phenotypically defined pMPP/CD90− cells were isolated by flow cytometry, and 2 × 103 pHSC/CD90low and 5 × 104 pMPP/CD90− cells were injected into sublethally irradiated NSG neonatal mice. Ten weeks after injection, BM was analyzed by flow cytometry for the expression of the indicated marker. Numbers within each plot indicate the percentage of cells in the indicated gates or quadrants. (E) Absolute numbers of CD14+, CD15+, and CD19+ cells recovered. (F) A linear correlation between the absolute numbers of CD45RA− cells recovered from the BM and the number of freshly sorted HSCs injected into mice is shown (R2 = 0.87). Absolute numbers of CD45RA− cells recovered from (C) and (D) were plotted on the linear graph and the corresponding number of HSCs injected was extrapolated. A total of 8 mice were analyzed.
In vivo potential of HSCs generated in vitro
Although phenotypically defined HSCs were maintained and expanded in the presence of Notch signals in vitro, the functional potential of these cells remained to be verified. We therefore compared the capacity of isolated DL4hi–derived day 14 (2 × 103) pHSCs with that of 25-fold more phenotypically defined MPP (CD90−) (pMPP) cells, isolated in parallel DL4hi–derived day 14 (5 × 104), to repopulate sublethally irradiated NOD/SCID/γcnull (NSG) mice.
After 10 weeks, BM samples from pHSC- and pMPP-reconstituted mice contained macrophages (CD33+CD14+), granulocytes (CD33+CD15+), and lymphoid cells (CD19+ B or CD7+ T cells) (Figure 2C-D). In all, 8 mice were analyzed in 2 groups of 4 (Figure 2E), and absolute numbers of myeloid or lymphoid cells were similar between the 2 groups. Therefore, both cultured pHSC and pMPP cells have lymphomyeloid reconstitution potential.
We next wanted to compare day 14 in vitro–grown pHSCs with freshly isolated HSCs for their ability to repopulate immune-deficient mice. NSG mice were injected with fresh HSCs ranging from 0.5 to 3 × 103 cells/mouse, and the resulting expansion of CD45RA− cells after 10 weeks within the BM of host mice was used to determine the expected cellular yield from injected HSCs (Figure 2F). A linear relationship was observed with the absolute number of donor CD45RA− cells in the BM correlating directly with the number of injected HSCs. We then extrapolated the number of CD45RA− cells recovered from mice injected with 2 × 103 day 14 in vitro–grown pHSCs and found that this corresponded to the equivalent of 0.5 × 103 freshly isolated HSCs, or a ∼0.25 times equivalency in cellular outcome (Figure 2F). In contrast, very few CD45RA− were found in mice injected with pMPP cells, even though ×25-fold more cells were injected. We conclude that pHSCs cultured in the presence of Notch signaling act as SCID-mouse repopulating cells (SRCs)27 similar to freshly isolated HSCs.
Effect of Notch inhibition on the maintenance/expansion of HSCs and/or CD45RA− subsets derived in vitro
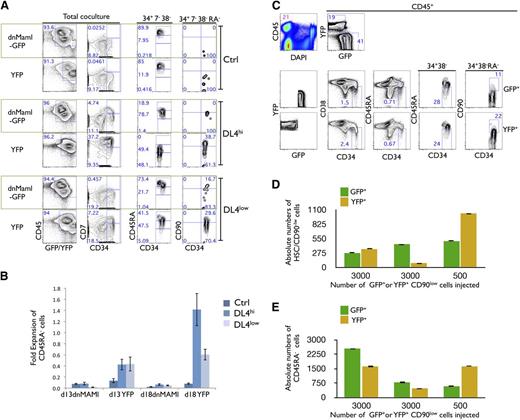
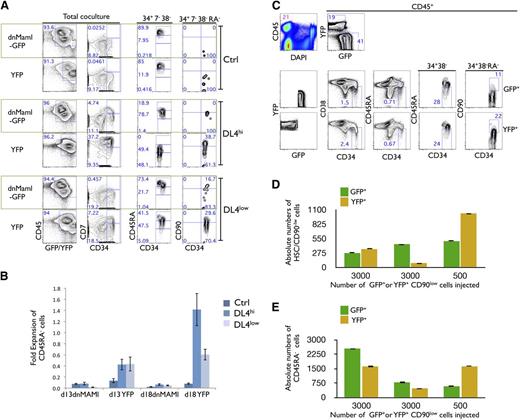
To test whether the in vitro expansion of pHSCs and/or CD45RA− cells is regulated by canonical Notch signaling, we transduced ex vivo HSCs with the dnMaml-GFP construct5 to prevent cells from responding to Notch signals. HSC cells expressing dnMaml-GFP and/or control yellow fluorescent protein (YFP) were generated and cultured on Ctrl, DL4hi, and DL4low cells for an additional 2 to 3 weeks. Analysis of CD34+CD38−CD7− gated cells from Ctrl cultures showed no CD45RA−, and therefore no CD90low cells, few CD45RAint, and a majority were CD45RAhi cells, regardless of the presence or the absence of dnMaml (Figure 3A last 2 columns). In sharp contrast, in CD34+CD38−CD7− gated cells derived from DL4hi cultures, expression of dnMaml-GFP totally abrogated the maintenance of both CD45RA− cells and CD90low cells, and accelerated the progression to the CD45RAint and CD45RAhi stages. Similar results were observed in DL4low cultures expressing the dnMaml-GFP construct.
In vitro, but not in vivo, expansion of HSCs is Notch dependent. (A) Freshly sorted HSCs were transduced with dnMaml-GFP or control YFP-only retroviral constructs, and sorted on day 6 posttransduction as GFP+ and YFP+ cells, cultured on OP9-Ctrl, -DL4hi -DL4low cells for 2 weeks. Cultures were analyzed by flow cytometry for the expression of the indicated markers as shown for total coculture. Cells gated as GFP+ or YFP+CD34+CD7−CD38− were further analyzed for the expression of CD45RA (34+7−38−). Additionally, CD45RA− gated cells were analyzed for the expression of CD90 (34+7−38−RA−). Numbers within each plot indicate the percentage of cells in the indicated gates or quadrants. Representative plots from at least 3 independent experiments are shown. (B) Cellular fold expansion of CD45RA− cells (5-10 × 103) is shown for the indicated coculture conditions at day 13 or 18 of culture relative to the start of coculture (ie, following the initial 7-day retroviral transduction period). Bar graphs show mean and SE of 4 independent experiments. (C) Freshly sorted HSCs were transduced with dnMaml-GFP or control YFP-only lentiviral constructs, and sorted on day 3 posttransduction as GFP+ or YFP+ CD34+CD7−CD38−CD90low (CD90low) cells. Equal numbers of transduced GFP+ or YFP+ CD90low cells were coinjected into sublethally irradiated NSG neonatal mice, and after 10 weeks, cells from the BM were analyzed by flow cytometry for the indicated markers. (C) Top rows indicate the frequency of donor cells in BM (CD45+) and expression of GFP and YFP is shown for CD45+-gated cells. Bottom rows show GFP+ and YFP+ gated cells as indicated. Numbers within each plot indicate the percentage of cells in the indicated gates or quadrants. (D) Absolute numbers of donor-derived CD90low cells and (E) CD45RA− cells from the BM of host mice, as in panel C. A representative experiment is shown, from 3 reconstituted mice.
In vitro, but not in vivo, expansion of HSCs is Notch dependent. (A) Freshly sorted HSCs were transduced with dnMaml-GFP or control YFP-only retroviral constructs, and sorted on day 6 posttransduction as GFP+ and YFP+ cells, cultured on OP9-Ctrl, -DL4hi -DL4low cells for 2 weeks. Cultures were analyzed by flow cytometry for the expression of the indicated markers as shown for total coculture. Cells gated as GFP+ or YFP+CD34+CD7−CD38− were further analyzed for the expression of CD45RA (34+7−38−). Additionally, CD45RA− gated cells were analyzed for the expression of CD90 (34+7−38−RA−). Numbers within each plot indicate the percentage of cells in the indicated gates or quadrants. Representative plots from at least 3 independent experiments are shown. (B) Cellular fold expansion of CD45RA− cells (5-10 × 103) is shown for the indicated coculture conditions at day 13 or 18 of culture relative to the start of coculture (ie, following the initial 7-day retroviral transduction period). Bar graphs show mean and SE of 4 independent experiments. (C) Freshly sorted HSCs were transduced with dnMaml-GFP or control YFP-only lentiviral constructs, and sorted on day 3 posttransduction as GFP+ or YFP+ CD34+CD7−CD38−CD90low (CD90low) cells. Equal numbers of transduced GFP+ or YFP+ CD90low cells were coinjected into sublethally irradiated NSG neonatal mice, and after 10 weeks, cells from the BM were analyzed by flow cytometry for the indicated markers. (C) Top rows indicate the frequency of donor cells in BM (CD45+) and expression of GFP and YFP is shown for CD45+-gated cells. Bottom rows show GFP+ and YFP+ gated cells as indicated. Numbers within each plot indicate the percentage of cells in the indicated gates or quadrants. (D) Absolute numbers of donor-derived CD90low cells and (E) CD45RA− cells from the BM of host mice, as in panel C. A representative experiment is shown, from 3 reconstituted mice.
These results point to Notch signaling as being necessary for the maintenance of CD45RA− and CD90low cells; in its absence, differentiation proceeds with the accelerated appearance of both CD45RAint and CD45RAhi stage cells. Additionally, cellular expansion of CD45RA− cells is dependent upon Notch signaling, as the fold expansion of YFP-only transduced cells relative to day 7 was greatest in DL4hi cocultures, while not being evident in dnMaml-Ctrl–transduced cells (Figure 2B).
Is Notch signaling required for the maintenance/expansion of HSCs in vivo?
We next tested whether this effect was also the case in vivo using competitive transplantation experiments. Equivalent numbers of sorted HSCs expressing either dnMaml-GFP+ or YFP+ were injected into sublethally irradiated neonatal NSG mice. Ten weeks later, the contribution of GFP- and/or YFP-expressing cells was analyzed in BM, thymus, and spleen. As shown in supplemental Figure 2A (left panels), the BM and spleen contained CD45+ donor cells, which included GFP+ and YFP+ cells. In both donor types, myeloid and B-lymphoid cells were similarly observed. Analysis of the BM from 10 independently reconstituted mice is shown in supplemental Figure 2D-G. Similar results were found in the spleen.
Analysis of the thymus showed CD45+ donor cells, but composed nearly entirely of YFP+ cells, with very few GFP+ cells (supplemental Figure 2B). The YFP+ cells included mainly T cells that expressed high levels of CD3. These findings clearly demonstrate effective abrogation of Notch signaling in GFP+ cells. To test the critical question of whether Notch signaling plays any role in the maintenance and/or self-renewal of human HSCs (SRC) in vivo, absolute numbers of donor-derived CD45RA− and pHSCs were compared in the BM, and were found similar in dnMaml-GFP and YFP-injected mice (Figure 3C-E). These results strongly suggest that Notch inhibition did not have an effect on the maintenance and/or self-renewal of SRCs within the BM microenvironment.
Effect of Notch engagement on the expansion of CD45RA subsets derived in vitro
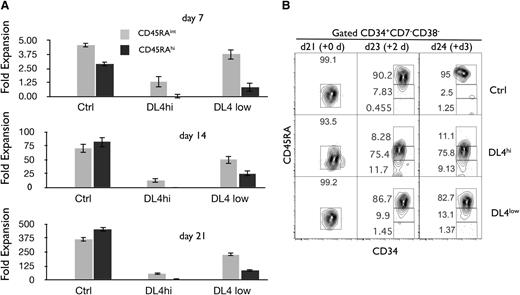
We next examined the consequences of the Notch-driven expansion of HSCs on early progenitors, namely the CD45RAint and CD45RAhi fraction (as shown in Figure 1C). We calculated the expansion of each progenitor fraction at weekly intervals based on the absolute number of cells recovered. Figure 4A shows that both CD45RAint and CD45RAhi fractions expanded the most in the absence of Notch signals (Ctrl). In the presence of Notch signaling, the generation of CD45RAint and CD45RAhi cells was not favored in particular with DL4hi (Figure 4 middle panels). These data suggest that a high or low level of Notch signaling expands CD45RA− and pHSCs, maintains them in an undifferentiated state, and restrains their differentiation into both CD45RAint and CD45RAhi progenitors.
Notch signals lead to an enhanced cellular expansion of CD45RA−cells and delay the cellular expansion of CD45RAintand CD45RAhiprogenitors. Cocultures were initiated with CD90low cells (HSCs) placed on OP9-Ctrl, -DL4hi, and -DL4low cells and analyzed by flow cytometry at days 7, 14, and 21 of culture. The cellular fold expansion based on the absolute numbers of cells recovered (from day 0 5-10 000 HSC plated) is shown for CD45RAint and CD45RAhi cells. (A) Bar graphs show mean and SE of at least 10 independent experiments. (B) Precursor-product relationship between CD45RAint and CD45RAhi cells. Cocultures were initiated with CD90low cells (HSCs) placed on OP9-Ctrl, -DL4hi, and -DL4low cells. After 21 days of culture, OP9-Ctrl, -DL4hi, and -DL4low cultures were harvested separately and CD45RA−/int cells were sorted by flow cytometry and recultured under the same conditions in stem media. This defines a new day 0 (purity analysis for this day 0 shown). Two and 3 days later, cells were analyzed for the expression of CD34 and CD45RA on gated CD34+CD38−CD7− cells. Numbers within each plot indicate the percentage of cells in the indicated gates. Representative plots of 3 independent experiments are shown.
Notch signals lead to an enhanced cellular expansion of CD45RA−cells and delay the cellular expansion of CD45RAintand CD45RAhiprogenitors. Cocultures were initiated with CD90low cells (HSCs) placed on OP9-Ctrl, -DL4hi, and -DL4low cells and analyzed by flow cytometry at days 7, 14, and 21 of culture. The cellular fold expansion based on the absolute numbers of cells recovered (from day 0 5-10 000 HSC plated) is shown for CD45RAint and CD45RAhi cells. (A) Bar graphs show mean and SE of at least 10 independent experiments. (B) Precursor-product relationship between CD45RAint and CD45RAhi cells. Cocultures were initiated with CD90low cells (HSCs) placed on OP9-Ctrl, -DL4hi, and -DL4low cells. After 21 days of culture, OP9-Ctrl, -DL4hi, and -DL4low cultures were harvested separately and CD45RA−/int cells were sorted by flow cytometry and recultured under the same conditions in stem media. This defines a new day 0 (purity analysis for this day 0 shown). Two and 3 days later, cells were analyzed for the expression of CD34 and CD45RA on gated CD34+CD38−CD7− cells. Numbers within each plot indicate the percentage of cells in the indicated gates. Representative plots of 3 independent experiments are shown.
Precursor-product relationship of CD45RA fractions derived in vitro
We next determined precursor-product relationships between CD45RAint and CD45RAhi fractions generated in vitro. CD45RAint were sorted from day 21 Ctrl, DL4hi, and DL4low cocultures and then recultured (new day 0) under the same conditions from which they were originally derived. As shown in Figure 4B, >80% of CD45RAint became CD45RAhi after only 2 days of further culture on Ctrl or DL4low, but cells progressed more slowly to CD45RAhi when recultured in the presence of a high Notch signal. This indicated that CD45RAint were precursors of CD45RAhi cells. We concluded that CD45RA− give rise to CD45RAint and then to CD45RAhi, with Notch signaling restraining this developmental progression.
Functional characterization of progenitor cells: bulk and clonal assays
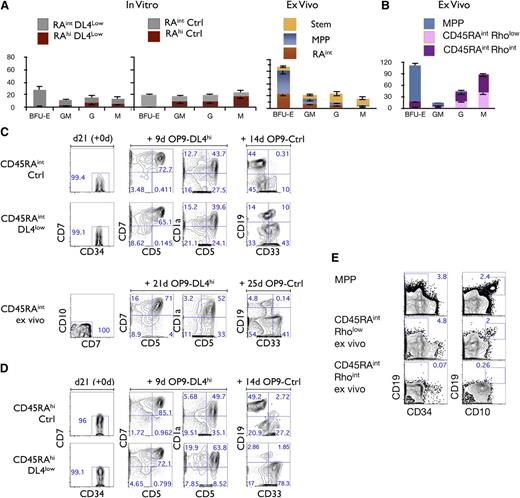
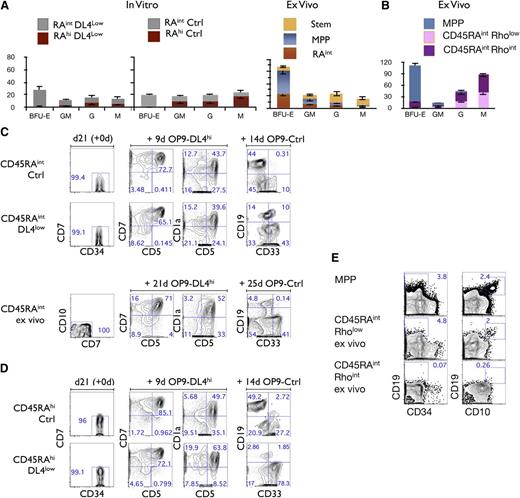
We next addressed the differentiation potential of CD45RAint and CD45RAhi subsets and examined their ability to generate colony-forming units (CFUs) in methylcellulose cultures. Figure 5A (top left panels) shows that CD45RAint cells derived from either Ctrl or DL4low cultures gave rise to BFU-E, GM, G, and M colonies. In striking contrast, very few if any BFU-E colonies were found in the CD45RAhi fraction from OP9-DL4low or -Ctrl cultures. Nevertheless, CD45RAhi cells have clear myeloid potential in that they gave rise to GM, G, and M colonies at higher frequencies than ex vivo MPP cells (Figure 5A). Therefore, CD45RAint and CD45RAhi cells both possess myeloid potential but CD45RAhi cells appear to have lost erythroid potential.
Lineage potential of CD45RAintand CD45RAhicells. (A) CFU-C generation from 500 plated, day 21–sorted CD45RAint and CD45RAhi cells, grown on OP9-DL4low (first panel) and/or OP9-Ctrl (second panel). CFU-C generation from ex vivo–derived Stem, MPP, and CD45RAintRholow/int CD133+CD7−CD10− (ex vivo CD45RAint) (third panel). Mean and SEM were calculated from 6 independent experiments. (B) CFU-C generation from 1000 plated ex vivo CD90− (MPP), CD45RAintRhoint cells or CD45RAintRholow cells. (C) Purity reanalysis of CD45RAint day 21–sorted fractions and ex vivo CD45RAint is shown as day 21 (+0d) (first column). Generation of CD5+, CD7+, and CD1a+ cells is shown in the middle columns and CD19+ cells vs CD33+ in the last column for CD45RAint derived from OP9-Ctrl and /or DL4low and ex vivo CD45RAint. (D) Purity reanalysis of CD45RAhi day 21–sorted fractions is shown as day 21 (+0d) (first column). Generation of T cells in vitro by CD45RAhi cells derived from OP9-Ctrl or OP9-DL4low cultures is shown in the middle columns, and generation of CD19+ or CD33+ cells is shown in the last column. Numbers within each plot indicate the percentage of cells in the indicated gates. This is a representative experiment of at least 10 independent experiments. (E) Ex vivo CD90− (MPP), CD45RAintRhoint cells or CD45RAintRholow cells were cultured on OP9-Ctrl for 26 days and analyzed for the expression of CD34, CD10, and CD19 by flow cytometry. Numbers within each plot indicate the percentage of cells in the indicated gates.
Lineage potential of CD45RAintand CD45RAhicells. (A) CFU-C generation from 500 plated, day 21–sorted CD45RAint and CD45RAhi cells, grown on OP9-DL4low (first panel) and/or OP9-Ctrl (second panel). CFU-C generation from ex vivo–derived Stem, MPP, and CD45RAintRholow/int CD133+CD7−CD10− (ex vivo CD45RAint) (third panel). Mean and SEM were calculated from 6 independent experiments. (B) CFU-C generation from 1000 plated ex vivo CD90− (MPP), CD45RAintRhoint cells or CD45RAintRholow cells. (C) Purity reanalysis of CD45RAint day 21–sorted fractions and ex vivo CD45RAint is shown as day 21 (+0d) (first column). Generation of CD5+, CD7+, and CD1a+ cells is shown in the middle columns and CD19+ cells vs CD33+ in the last column for CD45RAint derived from OP9-Ctrl and /or DL4low and ex vivo CD45RAint. (D) Purity reanalysis of CD45RAhi day 21–sorted fractions is shown as day 21 (+0d) (first column). Generation of T cells in vitro by CD45RAhi cells derived from OP9-Ctrl or OP9-DL4low cultures is shown in the middle columns, and generation of CD19+ or CD33+ cells is shown in the last column. Numbers within each plot indicate the percentage of cells in the indicated gates. This is a representative experiment of at least 10 independent experiments. (E) Ex vivo CD90− (MPP), CD45RAintRhoint cells or CD45RAintRholow cells were cultured on OP9-Ctrl for 26 days and analyzed for the expression of CD34, CD10, and CD19 by flow cytometry. Numbers within each plot indicate the percentage of cells in the indicated gates.
As shown in supplemental Figure 1A, a similar CD45RAint subset is present in CB cells, which are CD45RAintCD135+CD133+CD7−CD10−Rholow/int (ex vivo CD45RAint). By comparing the myeloid potential of in vitro–derived CD45RAint cells to ex vivo CD45RAint cells, we observed that both CD45RAint cells display erythromyeloid potential (Figure 5A third top panel). To further characterize the BFU-E potential within this population, we fractionated ex vivo CD45RAint cells into Rholow and Rhoint fractions as shown in supplemental Figure 1A and tested each fraction separately. Only the Rhoint fraction gave rise to BFU-E although both fractions generated GM, G, and M CFU colonies (Figure 5B).
The lymphoid potential of CD45RAint and CD45RAhi cells generated in vitro was also assessed. The ability to generate T and B cells was observed in both CD45RAint (Figure 5C) and CD45RAhi cells (Figure 5D). The comparison of in vitro CD45RAint to ex vivo CD45RAint revealed that ex vivo CD45RAint also had T- and B-lineage potential (Figure 5C). However, a lower frequency of B cells was consistently found as compared with in vitro–derived CD45RAint cells. When ex vivo–derived CD45RAintRholow/int cells were further fractionated into Rholow and Rhoint cells, almost no B cells were generated in the Rhoint fraction whereas the Rholow fraction did give rise to B cells (Figure 5E).
To define the potential of these fractions at the clonal level, we repeated these analyses under limiting dilution conditions. We found that every single clone of CD45RAint and CD45RAhi was able to generate T cells regardless of the presence or absence of Notch signaling during the first 3 weeks of culture. Interestingly, T cells were also generated from ex vivo CD45RAint clones at a similar frequency (Figure 5B; supplemental Figure 3A, supplemental Tables 1 and 2). Additionally, B cells, as well as macrophages and granulocytes (supplemental Figure 3, supplemental Tables 1 and 2), were also present at comparable frequencies from day 21 cocultures of clonally sorted CD45RAint and CD45RAhi cells.
Because the majority of CD45RAint or CD45RAhi clonal outcomes contained both CD19+ cells and CD33+ cells (supplemental Table 3) and given the T-cell frequencies observed (supplemental Table 2), our clonal analysis indicates that both CD45RAint and CD45RAhi cells were able to give rise to T, B, and myeloid cells. Therefore, a substantial fraction of both CD45RAint and CD45RAhi cells are lymphomyeloid precursors, the former compatible with an MPP.2 type of cell and the latter with a lymphoid-primed MPP (LMPP) type of cell.28
Ex vivo CD45RAintRholow cells have a similar potential to in vitro–generated CD45RAhi cells (supplemental Figure 5). Therefore, ex vivo–derived CD45RAintRholow cells are LMPP-like cells. However, ex vivo CD45RAintRhoint cells have the potential to generate at the clonal level GM, G, M, and T cells (supplemental Tables 1 and 2). Only a fraction of them also have in vitro erythroid potential. So, at least 2 subsets are present in ex vivo CD45RAint cells, a minority subset with erythromyeloid and lymphoid potential and a majority subset with myeloid and T-cell potential, but devoid of B and erythroid potential in vitro.
Molecular characterization of CD45RA subsets
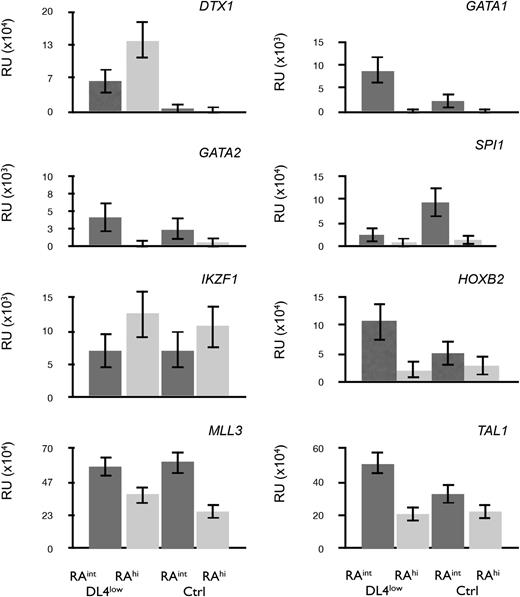
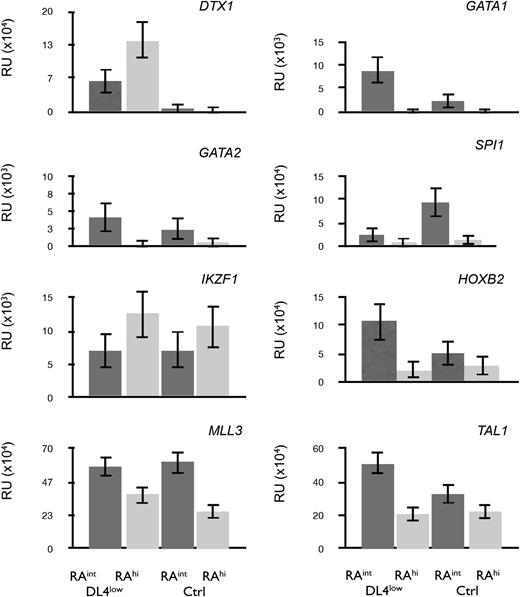
To further characterize the in vitro–generated CD45RAint and CD45RAhi cells, we performed a quantitative polymerase chain reaction (qPCR) analysis. Figure 6 shows that DTX1 (Deltex1) was expressed only on CD45RAint and CD45RAhi cells grown from DL4low cultures (Figure 6 top left panel) consistent with Notch signaling in these cultures. GATA1 and GATA2 transcripts were expressed in CD45RAint cells but were present at low levels in CD45RAhi cells. GATA1 expression was high in CD45RAint cells from DL4low cultures, which possessed a higher BFU-E frequency.
Profiling of CD45RAintand CD45RAhicells. qPCR for transcripts expressed in CD45RAint or CD45RAhi derived from OP9-DL4low or OP9-Ctrl and sorted on day 21. All samples were normalized to β-actin. Mean and SEM were calculated from 3 to 5 independent qPCR analyses.
Profiling of CD45RAintand CD45RAhicells. qPCR for transcripts expressed in CD45RAint or CD45RAhi derived from OP9-DL4low or OP9-Ctrl and sorted on day 21. All samples were normalized to β-actin. Mean and SEM were calculated from 3 to 5 independent qPCR analyses.
On the other hand, IKZF1 (Ikaros) transcripts were expressed at higher levels in CD45RAhi cells from either Ctrl or DL4low cultures. This, together with the low levels of GATA1, confirmed a similar pattern of expression to that of mouse cells with similar differentiation potential. Additionally, SPI1 (PU.1), HOXB2, MLL3, and TAL1 (SCL) (Figure 6) were expressed at higher levels in CD45RAint than in CD45RAhi cells from either Ctrl or DL4low cultures, correlating with their earlier stage of differentiation. Taken together, the gene expression patterns found in both lymphomyeloid progenitors generated in vitro correspond to the pattern of gene expression predicted from their differentiation potential.
In vivo potential of in vitro–generated CD45RAint and CD45RAhi and ex vivo CD45RAint cells
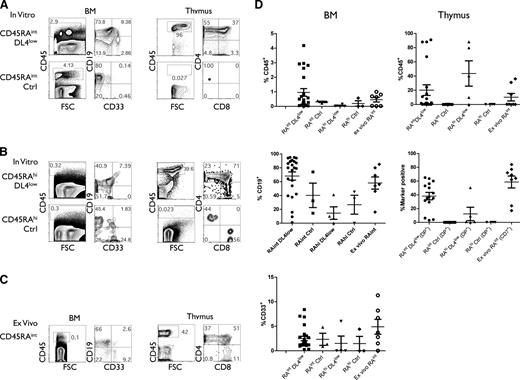
We next assessed the in vivo developmental potential of in vitro CD45RAint and CD45RAhi cells and ex vivo CD45RAint cells to reconstitute BM and thymi of BALB/c Rag−/−γcnull mice. A relatively low number (105) of CD45RAint and CD45RAhi cells from DL4low or Ctrl cultures (2-4 weeks) was injected into immune-deficient neonatal mice. CD45RAint and CD45RAhi cells isolated from DL4low cultures engrafted mice with donor CD45+ cells in the BM and thymus, including B and myeloid cells in the BM and T cells in the thymus, respectively, when analyzed 4 to 7 weeks postinjection (Figure 7A,B,D). We noted that different frequencies of CD4+CD8− cells, which are mainly immature single-positive cells, reflect the different temporal kinetics of reconstitution from the CD45RAint and CD45RAhi cells.
In vivo differentiation potential of CD45RA subsets. Sublethally irradiated immune-deficient neonates were injected with in vitro–derived or ex vivo–derived indicated cells at concentrations ranging from 0.1 to 1 × 105 cells per mouse. After 4 to 7 weeks, mice were sacrificed and thymi and/or BM cells were analyzed by flow cytometry for shown markers. (A,D) The percentage of donor CD45+ cells in BM and/or thymus from mice injected with CD45RAint derived from OP9-DL4low or OP9-Ctrl cultures (15 and 7 mice, respectively). (B,D) The percentage of CD45+ marker positive cells found in BM and/or thymus of mice injected with CD45RAhi derived from OP9-DL4low or OP9-Ctrl cultures (5 and 4 mice, respectively) or (C-D) ex vivo CD45RAint (7-8 mice).
In vivo differentiation potential of CD45RA subsets. Sublethally irradiated immune-deficient neonates were injected with in vitro–derived or ex vivo–derived indicated cells at concentrations ranging from 0.1 to 1 × 105 cells per mouse. After 4 to 7 weeks, mice were sacrificed and thymi and/or BM cells were analyzed by flow cytometry for shown markers. (A,D) The percentage of donor CD45+ cells in BM and/or thymus from mice injected with CD45RAint derived from OP9-DL4low or OP9-Ctrl cultures (15 and 7 mice, respectively). (B,D) The percentage of CD45+ marker positive cells found in BM and/or thymus of mice injected with CD45RAhi derived from OP9-DL4low or OP9-Ctrl cultures (5 and 4 mice, respectively) or (C-D) ex vivo CD45RAint (7-8 mice).
In contrast, mice injected with similar numbers of CD45RAint and CD45RAhi cells from Ctrl cultures were unable to effectively reconstitute the thymus, although they did reconstitute BM, with B and myeloid cells (Figure 7A-B). Notably, high levels of CD45+ engraftment were seen in the thymus of 15 mice injected with CD45RAint from DL4low cultures as compared with 7 mice injected with CD45RAint from Ctrl cultures (Figure 7D). Nonetheless, injection of higher numbers of these populations enabled CD45RAint and CD45RAhi cells from Ctrl cultures to reconstitute the thymus (data not shown). Thus, it appears that Notch signaling facilitated but was not required for thymic reconstitution. Lastly, ex vivo CD45RAint cells engrafted both the BM (B and myeloid cells) and thymus (Figure 7C). Results from 7 independent mice are presented in Figure 7D.
Discussion
We have defined culture conditions suitable for the maintenance of human HSCs, thus allowing us to directly test the effects of Notch signaling on prospectively defined HSCs. Here, we have found that HSCs are specifically affected by Notch signals in vitro, but do not rely on Notch signals for their expansion in vivo. In addition, our culture system enabled us to identify and characterize 2 new early progenitors, functionally akin to MPP-2 (CD45RAint) and LMPP (CD45RAhi) cells that were also found in CB.
The role of Notch on HSCs remains controversial. In aggregate, our current understanding points to a dichotomy in the role of Notch in HSC expansion: in vitro systems support a role for both mouse HSCs6 and human CD34+ cells4,7,8,10,29 while in vivo approaches discounted a role for mouse HSCs.5 Mechanisms of how Notch signaling influence the maintenance and expansion of human HSCs in vitro vs in vivo are complex and difficult to dissect because they require discrete fractionation of the different HSC subsets.4,30
As described by Anjos-Afonso,4 both the most primitive quiescent HSC (CD34−) and its progeny CD34+CD90lowCD49f+ express DTX1 and HES1, display different kinetics of reconstitution, and appear to be affected differentially upon Notch inhibition in vivo. Although a pharmacological inhibitor of Notch was used in this report, which is known to affect other nonhematopoietic tissues, maintenance and self-renewal of the CD34− subset was affected in vivo.4 However, here we show that competitive transplantation experiments using dnMaml for Notch inhibition in HSCs possessed equal reconstitution abilities in vivo, consistent with the results in mice.5 Nevertheless, whether loss of Notch signaling affected the temporal kinetics of reconstitution, as seen in mice,31 was not addressed.
Our results not only serve to extend the findings from Bernstein and colleagues,7 but more importantly, we now show that, in culture, HSCs are an important target of Notch signals. Additionally, it appears that cells at an early stage (CD34+CD38−CD7−CD10− CD45RA− cells [CD45RA−]) are maintained and expanded in the presence of Notch signals. Thus, by clarifying the target population exposed to Notch signals, a clearer picture of which cells are affected by this pathway has emerged.
A mechanism for how Notch signaling influences the maintenance and expansion of human HSCs in vitro is likely to involve several pathways regulated by Notch targets, such as cMyc, Cdkn1, and PTEN, all of which have been shown to affect HSC expansion/maintenance.32,33 However, the dichotomy between the requirements for Notch in vitro vs its dispensable role in vivo is likely due to the complex nature of the BM niche, in which Notch signals may be only one of many factors,17,34,35 while in vitro it becomes critically important. By removing HSCs from their normal BM niche, these cells would require a strong Notch signaling in vitro to compensate for the absence of the other signaling queues (supplemental Figure 4A).
We also showed that both in vitro–derived phenotypically defined HSC (CD45RA−CD90low) and phenotypically defined MPP (CD45RA−CD90−) populations were multipotent. However, upon transfer in to mice, in vitro–grown pHSCs were about 200 times more effective in generating CD45RA− cells than pMPPs, probably due to the fact that only CD90low cells were able to self-renew in vivo. In this regard, we found that in vitro–derived pHSCs approximated the SRC activity of freshly isolated HSCs when directly compared. Given the variability associated with in vivo manipulations, the use of a xenogeneic model system, and the relatively low numbers of cells injected, we can estimate that the 2 populations are likely similar in function.
The immediate stages following the differentiation of human CB HSCs are not fully appreciated, nor are the precursor-product relationships between these early progenitors clearly established. We now show that Notch signaling appears to delay the differentiation of CD45RA− cells in vitro, as the appearance of downstream CD45RAint and CD45RAhi cells is inhibited by Notch signals. We next characterized and showed that CD45RA− cells gave rise to CD45RAint, then to CD45RAhi and then to CD45RAhi (CD10+) cells (P.B., M.M., J.C.Z.-P., manuscript in preparation) in vitro, following a progression in which each fraction of lymphomyeloid progenitors corresponds to a more differentiated stage of development.
CD45RAint cells obtained in vitro were shown at both population and clonal levels, and at the transcript level, to be functionally equivalent to phenotypically defined multipotent cells (pMPP-2), whereas the CD45RAhi cells qualify as LMPP-like cells. Both subsets were able to reconstitute the same lymphomyeloid lineages when injected into immune-deficient mice. Interestingly, both subsets could reconstitute the thymus of immune-deficient mice, but when these in vitro cells had received Notch signals thymic reconstitution was greatly facilitated. In the absence of Notch signaling, thymic reconstitution occurs but only when higher numbers of cells are injected. So, Notch signals facilitate but are not required for thymic reconstitution. Similar functional subsets were characterized in CB (ex vivo CD45RAint). The presence of a weak erythroid differentiation potential in ex vivo Rhoint CD45RAint suggests that this phenotypically defined subset of CB cells could include both MPP-2 and GMP precursor cells.36,37 Interestingly, ex vivo CD45RAintRholow cells lacked erythroid potential, but had myeloid and lymphoid potential in vitro. Therefore, both subsets of ex vivo CD45RAint cells, Rhoint and Rholow, would appear to include cells that qualify as MPP-2 and LMPP, respectively.
Hao et al16 reported, at the clonal level, a Lin−CD34+CD7− early thymic lymphomyeloid precursor that has erythrolymphomyeloid potential, a finding unique to humans. The presence of thymic progenitors with such a potential correlates well with our in vitro–generated CD45RAint, as well as with ex vivo CD45RAint cells. Ex vivo CD45RAintRholow cells, described here as LMPP-like cells, are at a more immature stage of differentiation than the cells recently described by Kohn et al,18 in that they possess GM and G myeloid potential and express CD45RA at only intermediate levels. Haddad et al14 characterized in CB an ex vivo CD34+CD45RAhi cell, described as progeny of the in vitro–derived CD45RAint subset that shares characteristics with our in vitro–generated CD45RAhi subset.
The model presented in supplemental Figure 4B summarizes our findings. The human hematopoietic hierarchy appears similar to the murine system except for the fact that unlike the murine system, human pMPP-like cells with erythroid potential are able to reconstitute the thymus. We showed that the Notch-dependent maintenance of human pHSCs leads to an arrest in the differentiation of each of the lymphomyeloid progenitors (MPP-2 and LMPP) derived from it.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs Richard Miller and Norman Iscove for helpful discussion and critical review of the manuscript. We also thank Dr Els Verhoeyen for helpful technical advice on the generation and concentration of lentiviral supernatants. We acknowledge Courtney McIntosh for expertise in cell sorting, and Roxanne Holmes, John Xu, and Sanam Taheri for their technical support. We are grateful to Dr Rose Kung and staff of the Women and Babies program at Sunnybrook Health Sciences Centre for their ongoing support in providing us with umbilical CB.
This work was supported by grants to J.C.Z.-P. from the Terry Fox Foundation (TFF-15005), Canadian Institutes of Health Research (CIHR; MOP-119538 and HOP-83070), and The Krembil Foundation.
Authorship
Contribution: P.B. and P.S. performed and designed the research; M.M. and D.D. generated the OP9-DL4 lines; E.H. provided the umbilical CB samples; G.K. performed the flow cytometric cell sorts; and P.B. and J.C.Z.-P. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Juan Carlos Zúñiga-Pflücker, Department of Immunology, University of Toronto, Sunnybrook Research Institute, 2075 Bayview Ave, Toronto, ON, M4N 3M5, Canada; e-mail: jczp@sri.utoronto.ca.