Key Points
CD25 is a predictive biomarker for sensitivity to PIM inhibitors in AML cells.
PIM inhibitors may prolong overall/relapse-free survival through attenuating STAT5 activation and destabilizing MYC in CD25+ AML cells.
Abstract
Postchemotherapy relapse presents a major unmet medical need in acute myeloid leukemia (AML), where treatment options are limited. CD25 is a leukemic stem cell marker and a conspicuous prognostic marker for overall/relapse-free survival in AML. Rare occurrence of genetic alterations among PIM family members imposes a substantial hurdle in formulating a compelling patient stratification strategy for the clinical development of selective PIM inhibitors in cancer. Here we show that CD25, a bona fide STAT5 regulated gene, is a mechanistically relevant predictive biomarker for sensitivity to PIM kinase inhibitors. Alone or in combination with tyrosine kinase inhibitors, PIM inhibitors can suppress STAT5 activation and significantly shorten the half-life of MYC to achieve substantial growth inhibition of high CD25-expressing AML cells. Our results highlight the importance of STAT5 and MYC in rendering cancer cells sensitive to PIM inhibitors. Because the presence of a CD25-positive subpopulation in leukemic blasts correlates with poor overall or relapse-free survival, our data suggest that a combination of PIM inhibitors with chemotherapy and tyrosine kinase inhibitors could improve long-term therapeutic outcomes in CD25-positive AML.
Introduction
In acute myeloid leukemia (AML), treatment options for relapsed patients are limited and, therefore, remain a significant unmet medical need. It is believed that a population of chemoresistant leukemic stem cells (LSCs) is at least partially responsible for relapse after chemotherapy.1 Efforts to uncover LSC markers have revealed CD25 (IL2RA) as a potential candidate.2 Furthermore, retrospective studies have demonstrated that patients with a higher percentage of CD25-positive blasts manifest poor overall survival or relapse-free survival.3-5 Because CD25 is a well-established STAT5-regulated gene, high CD25 expression could likely be a result of hyperactivation of STAT5.6
STAT5 transcription factors, represented by isoforms STAT5A and STAT5B, are known to be crucial in both normal hematopoiesis and hematological malignancies.7 The extent of STAT5 activation depends on the interplay of multiple intra- and intermolecular events, including tyrosine phosphorylation, serine/threonine phosphorylation, and dimer/tetramer formation.8 STAT5 tyrosine phosphorylation events are usually regulated by intracellular or receptor tyrosine kinases.9-12 However, suppression of STAT activity merely through inhibition of upstream tyrosine kinases may lead to compensatory mechanisms that reactivate downstream signaling pathways.13
The PIM kinases are believed to be constitutively active and transcriptionally regulated at least in part by the JAK-STAT signaling pathway.12 Whether functional interactions among STAT transcription factors and the PIM kinase family extend beyond this linear paradigm has not been fully explored. Nevertheless, the fact that STAT5 activation induces interleukin 6 gene transcription, and that PIM2 has been shown to be required for interleukin 6 expression, implies that a PIM family-mediated feed-forward mechanism regulating STAT activity may exist.14,15
The PIM serine/threonine kinase family is composed of 3 highly homologous proteins: PIM1, PIM2, and PIM3. Functional redundancy of the 3 PIM kinases has previously been demonstrated both in vitro and in vivo.16-18 Furthermore, it is only in the absence of all 3 PIM kinases that knockout mice display overt reduced body size and impaired response to hematopoietic growth factors, although the animals remain viable and fertile.19 Given the functional redundancy of the 3 PIM kinases, a pan-PIM inhibitor will likely be required to achieve maximal efficacy as a targeted anticancer agent.
The PIM kinases mediate their activity through phosphorylation of a wide range of cellular substrates belonging to a number of crucial signaling pathways, including, among others, MYC, BAD, S6, and 4EBP1.20-23 Functional interactions between the PIM family and MYC have been most well characterized in genetic rodent models of lymphomagenesis.17 Employing biochemical and overexpression methodologies, PIM kinases have been shown to regulate MYC protein stability through direct phosphorylation of MYC S329.20 However, these findings do not preclude the possibility that there are additional regulatory mechanisms that the PIM family uses to regulate endogenous MYC activity.
MYC, STAT5, and PIM kinases have all been demonstrated to play critical roles in the maintenance, expansion, and self-renewal of hematopoietic stem cells.24-26 Given overlapping features between hematopoietic stem cells and LSCs, it is not surprising that STAT5 and MYC also play significant roles in leukemia-initiating activity.27,28 Simultaneous suppression of STAT5 and MYC activation may therefore negatively regulate the expansion of the LSC population.
In the present study, we uncovered the mechanism rendering a subset of AML cell lines sensitive to PIM inhibitors and solidified the role of CD25 as a mechanistically relevant predictive biomarker for PIM kinase inhibition. In particular, we reveal causal functional interactions among PIM kinases, STAT5, and MYC in high CD25-expressing cells. Our results may provide a hypothesis-driven endpoint for clinical trial design.
Methods
Detailed methods are available in the supplemental Methods and Figures, available on the Blood Web site. Microarray data available at Gene Expression Omnibus (GEO), accession number GSE59808.
Results
Identification of AML cell lines sensitive to the inhibition of PIM kinases
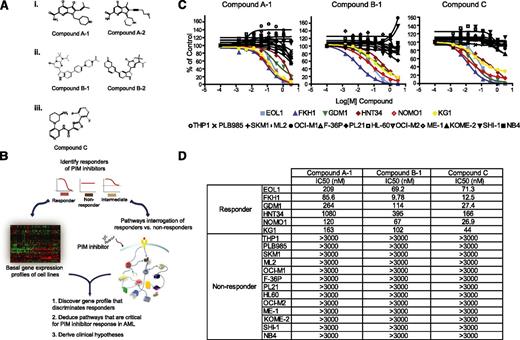
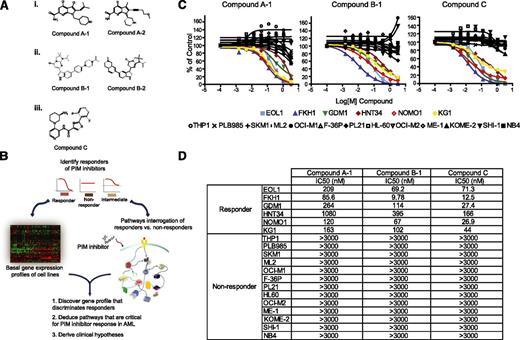
To characterize the underlying mechanisms of AML cell line sensitivity to PIM kinase inhibition, we employed an expanded panel of 29 commercially available AML cell lines to initiate responder studies. To ensure that the results obtained were a result of the on-target activity of compounds, 5 PIM inhibitors representing 3 diverse scaffolds were used throughout this report (Figure 1A). All 5 PIM inhibitors demonstrated significant potency against PIM1/2/3, as well as a high degree of selectivity against a panel of 192 kinases (supplemental Table 1). As depicted in Figure 1B, AML cell lines were treated with 3 structurally diverse PIM inhibitors (compound A-1, compound B-1, and compound C29,30 ) and subsequently categorized as responders, nonresponders, or intermediates, based on the extent of their proliferative response to compounds. In parallel, the basal genomewide messenger RNA (mRNA) expression profile of each cell line was obtained and a subset of responders and nonresponders was further subjected to mechanistic studies, using 3 structurally diverse PIM inhibitors, with a focus on evaluating components of the signaling pathways PIM kinases are known to mediate.
A subset of AML cell lines is sensitive to PIM kinase inhibitors. (A) Structures of 5 PIM kinase inhibitors representing 3 unique chemical scaffolds. (B) Schematic of the strategy employed to identify and molecularly characterize responders to PIM kinase inhibition. (C) The specified AML cell lines were treated with 3 diverse scaffolds of PIM kinase inhibitors at 8 concentrations (1:3 serial dilutions with maximal concentration of 3 µM). The distinctive sensitivity of a subset of AML cell lines to the impediment of PIM kinase activity is consistent across all 3 compounds tested, indicative of the on-target cellular response. (D) Summary of IC50 values of each PIM inhibitor used across responders and nonresponders.
A subset of AML cell lines is sensitive to PIM kinase inhibitors. (A) Structures of 5 PIM kinase inhibitors representing 3 unique chemical scaffolds. (B) Schematic of the strategy employed to identify and molecularly characterize responders to PIM kinase inhibition. (C) The specified AML cell lines were treated with 3 diverse scaffolds of PIM kinase inhibitors at 8 concentrations (1:3 serial dilutions with maximal concentration of 3 µM). The distinctive sensitivity of a subset of AML cell lines to the impediment of PIM kinase activity is consistent across all 3 compounds tested, indicative of the on-target cellular response. (D) Summary of IC50 values of each PIM inhibitor used across responders and nonresponders.
Of the 29 AML cell lines tested, 6 were found to be sensitive to PIM kinase inhibition, and 13 were found to be insensitive. These findings were consistent across all 3 PIM inhibitors employed (Figure 1C). The 50% inhibitory dose (IC50) values obtained with these compounds in the cell proliferation assays were shown in Figure 1D. Of note, similar results were obtained with EOL1, KG1, OCI-M1, HL60, and OCI-M2 cell lines in a recent publication using AZD1208, another pan-PIM kinase inhibitor.31 The remaining 10 cell lines did not consistently achieve more than 70% inhibition at the highest concentration (3 µM) across the 3 PIM inhibitors tested and were categorized as intermediates (supplemental Figure 1). For all subsequent analyses, we intentionally focused exclusively on responder and nonresponder cell lines to maximize the chance of acquiring robust biological signals.
Subset of genes from the responder expression signature is associated with poor survival in AML patients
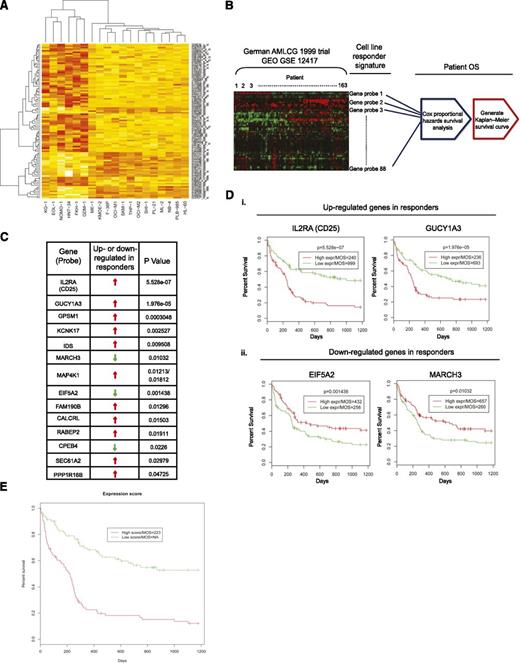
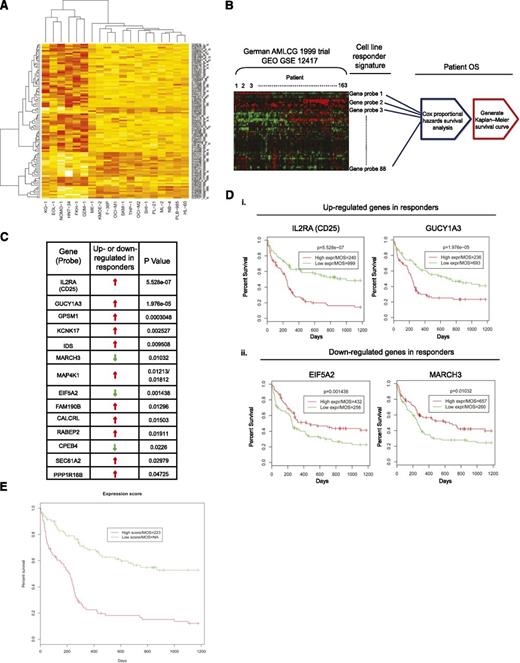
To derive a responder gene expression signature for PIM kinase inhibition, linear model analysis was performed to identify significant probes that differentiate responders and nonresponders. We applied multiple testing correction to the raw P values to control the false-discovery rate; however, it turned out to be too stringent, given the small sample size in this study (responders, n = 6; nonresponders, n = 13). We therefore selected the top 100 probes that had the smallest P values, which represented 81 different genes (Figure 2A; supplemental Table 2; supplemental Figure 2; supplemental Methods). To investigate the generalizability and the clinical relevance of the signature, we used the expression profiles of 163 patients in the German AML Cooperative Group (AMLCG) 1999 trial and the associated patient survival time as our test data. A total of 88 probes of 100 responder probes were found in the test data and were used in the subsequent analysis. To assess the significance of clinical relevance of each individual candidate gene in the responder expression signature, Cox proportional hazard survival analysis was performed for each probe in the test data (Figure 2B). Using a cutoff of P < .05, we identified a group of 14 genes that were able to predict clinical outcomes according to their expression levels (Figure 2C). Strikingly, the mRNA expression levels of CD25 and GUCY1A3, both of which have previously been reported to be significantly elevated in LSCs (supplemental Table 2),2,32,33 were shown to discriminate overall patient survival with superior statistical significance (Figure 2D; supplemental Figure 3 for all Kaplan-Meier survival curves). Specifically, patients with high CD25 expression had a median overall survival (MOS) of 240 days, in contrast to 999 days for patients with low expression of CD25 (Figure 2Di, CD25). Similarly, high expression of GUCY1A3 resulted in a MOS of 236 days, whereas low expression led to a MOS of 693 days (Figure 2Di, GUCY1A3). The mRNA expression levels of both EIF5A2 and MARCH3 were shown to be downregulated in responders. Although the expression levels of these 2 genes did not distinguish overall survival as prominently as those of CD25 and GUCY1A3, the fact that the associated overall survival curves are the inverse of those in the “upregulated genes” category further supports the notion that a subset of the PIM inhibitor responder signature identified correlates with poor clinical outcome in AML (Figure 2Dii, EIF5A2 and MARCH3). Further, we fit a Cox model using glmnet package in R.2.14.1,34 with lasso penalty (α = 1). The fitted model was cross-validated, and 10 probes were kept in the model. Not surprisingly, CD25 was one of the genes represented by the probes with a regression coefficient equal to 0.34, suggesting that higher expression of CD25 may lead to poorer overall survival (see supplemental Table 3 for coefficients of 10 probes). We then calculated a combined gene expression score for each patient, using the fitted values predicted by the glmnet model. We found that those patients with a high expression score exhibited a poorer overall survival (Figure 2E; P = 2.71e−7).
Statistical analysis for identification of responder signature. (A) Heat map of top 100 probes that significantly differentiate responders (KG1, EOL-1, NOMO-1, HNT-34, FKH-1, and GDM-1) from nonresponders. Red, high expression; yellow, low expression. (B) Workflow of the statistical validation using German AMLCG 1999 clinical trial data as test data. The top 100 responder probes identified from the cell line study were used to retrieve relevant information from the test data. Using patient overall survival (OS) time, Cox proportional hazards survival analysis and Kaplan-Meier analysis were performed. (C) Fourteen genes in the test data were found to be significantly associated with patient OS time. (D) High expression of CD25 or GUCY1A3 (i) and low expression of EIF5A2 or MARCH3 (ii) were found to be significantly associated with shorter overall survival time. MOS, median of overall survival time. (E) Kaplan-Meier plot shows the high expression score from the combined signature significantly predicted poor patient survival time in the German AMLCG 1999 clinical trial data (P = 2.71e−7). MOS of “NA” indicates that more than 50% of the patients in the “low score” group survived until the end of the study.
Statistical analysis for identification of responder signature. (A) Heat map of top 100 probes that significantly differentiate responders (KG1, EOL-1, NOMO-1, HNT-34, FKH-1, and GDM-1) from nonresponders. Red, high expression; yellow, low expression. (B) Workflow of the statistical validation using German AMLCG 1999 clinical trial data as test data. The top 100 responder probes identified from the cell line study were used to retrieve relevant information from the test data. Using patient overall survival (OS) time, Cox proportional hazards survival analysis and Kaplan-Meier analysis were performed. (C) Fourteen genes in the test data were found to be significantly associated with patient OS time. (D) High expression of CD25 or GUCY1A3 (i) and low expression of EIF5A2 or MARCH3 (ii) were found to be significantly associated with shorter overall survival time. MOS, median of overall survival time. (E) Kaplan-Meier plot shows the high expression score from the combined signature significantly predicted poor patient survival time in the German AMLCG 1999 clinical trial data (P = 2.71e−7). MOS of “NA” indicates that more than 50% of the patients in the “low score” group survived until the end of the study.
High CD25 expression represents STAT5 activation and can be suppressed through inhibition of PIM kinase activity
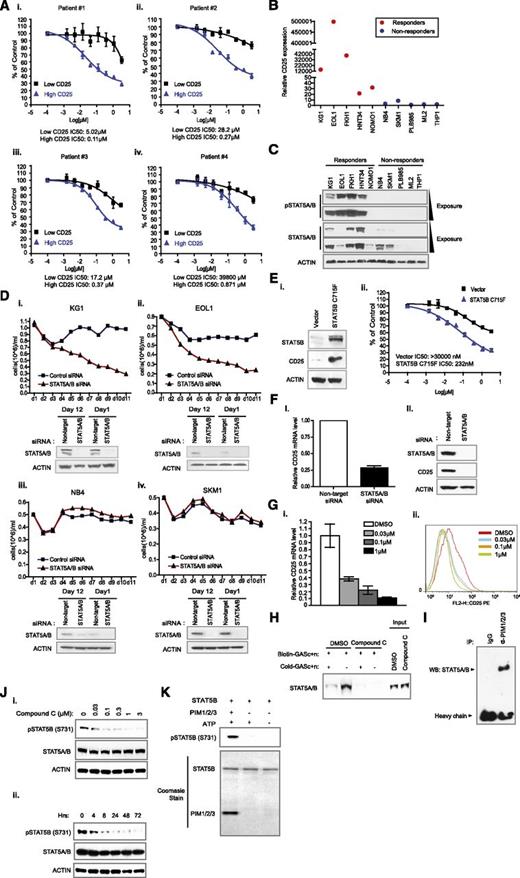
CD25 has been reported to be highly expressed in chemotherapy-resistant, quiescent LSCs.2 In several retrospective studies investigating CD25 as a prognostic marker in AML using an immunophenotyping approach, a higher percentage of CD25-positive cells (>10% or >20%) in leukemic blasts were found to correlate with poor overall and relapse-free survival.3-5 Here we demonstrated that a higher mRNA expression level of CD25 also correlates with poor overall survival (Figure 2Di, CD25). Because CD25 is a prominent candidate gene in the responder signature for PIM inhibitors with potential clinical relevance, we wanted to assess whether subpopulations of primary leukemic blasts from patients with differential expression of CD25 display unique response to PIM inhibitors. As shown in Figure 3A, using 4 patient samples (>75% blasts) with a CD25-positive cell population, we found that the subpopulation with high CD25 expression is exquisitely sensitive to compound C compared with the cells with low CD25 expression (supplemental Figure 4 for sorting results; supplemental Table 4 for patient information). Because of this observation and its clinical significance, we focused on CD25 for our subsequent studies.
Inhibition of PIM kinase activity suppresses growth of AML cells with high CD25 expression by down-regulating STAT5 activity. (A) Differential response of patient leukemia blasts subpopulations to the inhibition of PIM kinase activity. Primary leukemia blasts from 4 patients were sorted into high-CD25 expression or low-CD25 expression subpopulations and treated with the indicated concentrations of PIM inhibitor. IC50 values were specified. (B) Verification of CD25 mRNA levels in responder cell lines. Quantitative reverse-transcription polymerase chain reaction assays measuring CD25 mRNA levels in the indicated cell lines were implemented. Relative CD25 expression was calculated by normalizing the CD25 mRNA expression value of each cell line to that of the PLB985 cell line. (C) Level of STAT5 tyrosine phosphorylation in a panel of responders and nonresponders. Responders of PIM kinase inhibition exhibited a high level of STAT5 tyrosine phosphorylation (Y694/699), with the exception of NOMO1. In some nonresponders, the expression of total STAT5 was not obvious. (D) Effect of STAT5 knockdown on the growth of KG1, EOL1, NB4, and SKM1 cell lines. Sustained knockdown of endogenous STAT5A/B was achieved by continuous exposure to Accell siRNAs (see supplemental Methods) over the course of specified experiments. Culture volume was diluted 1:1 every day to avoid overconfluency and ensure the optimal growth of cells. (E) Proliferative response to PIM kinase inhibition on expression of constitutive STAT5B in a nonresponder cell line. Introduction of STAT5B C715F drastically upregulated CD25 levels in a nonresponder ML2 cell line (i). (F) Regulation of CD25 expression in KG1 cells. Knockdown of STAT5 in KG1 cells abrogated CD25 mRNA (i) and protein expression (ii). (G) The effect of PIM kinase inhibition on CD25 mRNA expression. PIM inhibitor dose-dependently downregulates CD25 expression at the mRNA level (i) and the protein level (ii). (H) Modulation of STAT5 DNA binding capacity in the presence of a PIM inhibitor. Nuclear extracts were made from either dimethylsulfoxide or compound C-treated cells and incubated with biotin-labeled double-stranded oligonucleotides derived from a CD25 promoter region that contains STAT5 DNA binding sequences (Biotin-GASc+n). “Cold- GASc+n” denotes unlabeled oligonucleotide that serves as a control for binding specificity. (I) Interaction between PIM kinase family and STAT5 transcription factors. Immunoprecipitation of PIM kinases was carried out in radioimmunoprecipitation assay buffer, and the presence of STAT5 was detected by western blot. (J) Modulation of STAT5B pS731 by PIM kinase inhibition. Treatment with PIM inhibitor significantly downregulated S731 phosphorylation on STAT5B in a dose-dependent (i) and time-dependent (ii) manner. (K) Direct phosphorylation of STAT5B S731 by PIM kinases. Biochemical reaction was carried out in the presence of recombinant STAT5B and PIM kinases. Protein input is illustrated by Coomassie staining (lower).
Inhibition of PIM kinase activity suppresses growth of AML cells with high CD25 expression by down-regulating STAT5 activity. (A) Differential response of patient leukemia blasts subpopulations to the inhibition of PIM kinase activity. Primary leukemia blasts from 4 patients were sorted into high-CD25 expression or low-CD25 expression subpopulations and treated with the indicated concentrations of PIM inhibitor. IC50 values were specified. (B) Verification of CD25 mRNA levels in responder cell lines. Quantitative reverse-transcription polymerase chain reaction assays measuring CD25 mRNA levels in the indicated cell lines were implemented. Relative CD25 expression was calculated by normalizing the CD25 mRNA expression value of each cell line to that of the PLB985 cell line. (C) Level of STAT5 tyrosine phosphorylation in a panel of responders and nonresponders. Responders of PIM kinase inhibition exhibited a high level of STAT5 tyrosine phosphorylation (Y694/699), with the exception of NOMO1. In some nonresponders, the expression of total STAT5 was not obvious. (D) Effect of STAT5 knockdown on the growth of KG1, EOL1, NB4, and SKM1 cell lines. Sustained knockdown of endogenous STAT5A/B was achieved by continuous exposure to Accell siRNAs (see supplemental Methods) over the course of specified experiments. Culture volume was diluted 1:1 every day to avoid overconfluency and ensure the optimal growth of cells. (E) Proliferative response to PIM kinase inhibition on expression of constitutive STAT5B in a nonresponder cell line. Introduction of STAT5B C715F drastically upregulated CD25 levels in a nonresponder ML2 cell line (i). (F) Regulation of CD25 expression in KG1 cells. Knockdown of STAT5 in KG1 cells abrogated CD25 mRNA (i) and protein expression (ii). (G) The effect of PIM kinase inhibition on CD25 mRNA expression. PIM inhibitor dose-dependently downregulates CD25 expression at the mRNA level (i) and the protein level (ii). (H) Modulation of STAT5 DNA binding capacity in the presence of a PIM inhibitor. Nuclear extracts were made from either dimethylsulfoxide or compound C-treated cells and incubated with biotin-labeled double-stranded oligonucleotides derived from a CD25 promoter region that contains STAT5 DNA binding sequences (Biotin-GASc+n). “Cold- GASc+n” denotes unlabeled oligonucleotide that serves as a control for binding specificity. (I) Interaction between PIM kinase family and STAT5 transcription factors. Immunoprecipitation of PIM kinases was carried out in radioimmunoprecipitation assay buffer, and the presence of STAT5 was detected by western blot. (J) Modulation of STAT5B pS731 by PIM kinase inhibition. Treatment with PIM inhibitor significantly downregulated S731 phosphorylation on STAT5B in a dose-dependent (i) and time-dependent (ii) manner. (K) Direct phosphorylation of STAT5B S731 by PIM kinases. Biochemical reaction was carried out in the presence of recombinant STAT5B and PIM kinases. Protein input is illustrated by Coomassie staining (lower).
To further validate the gene expression data, we measured CD25 expression, using real-time quantitative polymerase chain reaction. As shown in Figure 3B and supplemental Figure 5, CD25 mRNA is expressed at higher levels in responders compared with all nonresponders tested, with KG1, EOL1, and FKH1 cell lines having particularly high expression of CD25. It is well-established that CD25 is transcriptionally regulated by STAT5.6 We therefore assessed STAT5 activation status in both responder and nonresponder cell lines. As shown in Figure 3C and supplemental Figure 6, pSTAT5 (Y694/699) levels in responders were strikingly higher than those in nonresponders, with the exception of the NOMO1 cell line. Interestingly, some of the nonresponders did not appear to express detectable STAT5A/B (Figure 3C). To assess the functional interaction between PIM kinases and STAT5, we employed small interference RNA (siRNA) to deplete expression of STAT5A/B in KG1, EOL1 (both STAT5-positive responders), NB4, and SKM1 (both STAT5-positive nonresponders) cell lines and examined their growth rates. As shown in Figure 3D, STAT5A/B knockdown only inhibited the growth of KG1 and EOL1, not that of NB4 and SKM1 cell lines. When constitutively active STAT5B (STAT5B S715F)35 was introduced into a nonresponder ML2 cell line with little or no expression of STAT5, its expression resulted in an increased level of CD25 (Figure 3Ei) and rendered the cells more sensitive to compound C (Figure 3Eii).35 These results suggest that activation of STAT5 is a crucial mechanism that confers sensitivity to PIM kinase inhibition.
The expression of the PIM kinases is regulated by STAT5 transcription factor.12 However, PIM2 has been shown to be required for interleukin 6 expression, which is regulated at least in part by STAT5.14,15 These data suggest that PIM kinases may augment STAT5 activity through a feed-forward mechanism to maximally amplify the STAT5 proliferative signal. In the responder KG1 cell line, STAT5A/B is required for regulating CD25 mRNA expression, as knockdown of STAT5A/B significantly repressed the mRNA levels of CD25 (Figure 3Fi) and resulted in a marked decrease of CD25 protein levels (Figure 3Fii). Using CD25 as a surrogate marker for STAT5 activity, we investigated whether PIM inhibition could affect STAT5 activity. As demonstrated in Figure 3G, the PIM inhibitor compound C dose-dependently suppressed CD25 mRNA expression (Figure 3Gi), which translated into a significant reduction of CD25 functional protein expression in KG1 cells, as demonstrated by FACS analysis (Figure 3Gii). The repression of CD25 mRNA expression by PIM kinase inhibition is likely mediated through the disruption of STAT5-DNA interaction, as treatment with compound C severely reduced the binding capacity of STAT5A/B to the CD25 promoter oligo containing STAT5 binding elements (Figure 3H).36 We next evaluated whether a physical association, either direct or indirect, exists between PIM kinases and STAT5A/B transcription factors. The interaction between PIM kinases and STAT5A/B appeared to be robust, as immunoprecipitation of PIM1/2/3 in high-stringency radioimmunoprecipitation assay buffer allowed pull-down of a protein complex containing STAT5A/B (Figure 3I).
In addition to tyrosine phosphorylation, it is known that serine/threonine phosphorylation and dimer/tetramer formation contribute to the regulation of STAT5 activity.8,35 To investigate modulation of cellular STAT5A/B serine/threonine phosphorylation by PIM kinases, we evaluated phosphorylation of C-terminal serine/threonine residues that are known to regulate STAT5A/B activity with available antibodies. We found that phosphorylation of STAT5B on S731 could be significantly downregulated in a dose-dependent (Figure 3Ji) and time-dependent manner (Figure 3Jii) by compound C. Furthermore, in a well-defined in vitro biochemical setting, recombinant PIM kinases are capable of directly phosphorylating recombinant STAT5B on S731 (Figure 3K). Serine phosphorylation of STAT5B at S731 was previously demonstrated to be induced after growth factor stimulation and to regulate STAT5B activity in conjunction with tyrosine phosphorylation.37 Curiously, substantial inhibition of C-terminal STAT5 tyrosine phosphorylation was only observed at a higher dose (1 µM) of compound treatment and at a later point (supplemental Figure 7), suggesting PIM kinases play a role in facilitating STAT5 C-terminal serine phosphorylation and, possibly, in amplifying STAT5 signaling through a positive-feedback mechanism.
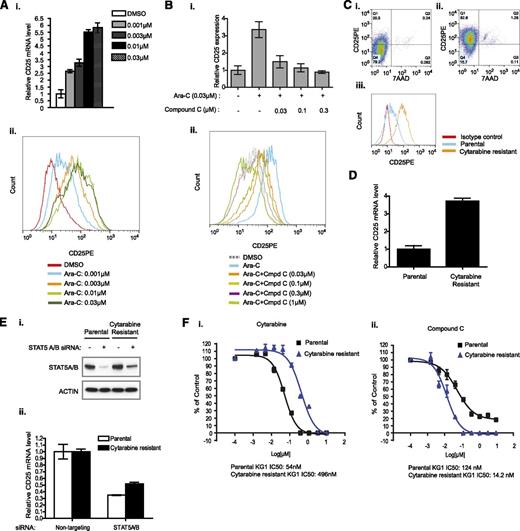
Cytarabine-resistant KG1 cell line expresses higher CD25 and is more sensitive to PIM kinase inhibition than parental KG1
Retrospective studies have suggested that a preexisting CD25-positive population in leukemic blasts may contribute to the development of relapse subsequent to standard-of-care chemotherapy.3-5 A similar study also showed that high STAT5 activity corresponds to poorer overall and relapse-free survival in AML.38 In addition, the evaluation of CD25-positive populations prior, during, and after induction chemotherapy showed that a majority of poorly responding patients demonstrated an increased percentage of CD25-positive cells at day 7 and day 21 after initiation of induction chemotherapy.39 These clinical results suggest the possibility that chemotherapy may enhance the expression of CD25 in a subset of cells through activation of STAT5 activity and contribute to the subsequent relapse of disease. To determine whether this observation could be recapitulated in an in vitro system, we treated KG1 cells with sub-IC50 doses of cytarabine and monitored CD25 mRNA expression levels (supplemental Figure 8). Cytarabine dose-dependently increased both CD25 mRNA (Figure 4Ai) and functional protein expression, as determined by fluorescence-activated cell sorter analysis (Figure 4Aii). Interestingly, the upregulation of CD25 mRNA and protein expression by cytarabine could be overcome by treatment with compound C in a dose-dependent manner (Figure 4B). Using an IC50 concentration of 30 nM, we established a cytarabine-resistant KG1 cell line in vitro. Subsequent characterization of these cytarabine-resistant KG1 cells revealed a marked increase of a CD25-positive population, functional CD25 protein expression, and CD25 mRNA compared with the parental KG1 cell line (Figure 4Ci-iii and Figure 4D). Importantly, STAT5 was still required for CD25 mRNA expression in the cytarabine-resistant line, as knockdown of STAT5A/B in cytarabine-resistant cells still inhibited the CD25 mRNA level significantly (Figure 4E). Previously, we hypothesized that inhibition of PIM kinase activity could diminish the activity of STAT5, thereby leading to inhibition of cell proliferation. This would suggest that the cytarabine-resistant line should be more sensitive to PIM kinase inhibition. As predicted, the cytarabine-resistant KG1 line exhibited a ∼9-fold increased IC50 value compared with the parental KG1 cell line in the presence of cytarabine (Figure 4Fi). In contrast, the same cytarabine-resistant KG1 cells displayed increased sensitivity to compound C compared with the parental line (Figure 4Fii).
Cytarabine-resistant cells express higher level of CD25 and are more sensitive to PIM kinase inhibition. (A) The effect of cytarabine on CD25 expression in cells. KG1 cells were treated with cytarabine at the indicated concentrations. The expression of CD25 was examined at the mRNA (i) and protein (ii) levels by quantitative reverse-transcription polymerase chain reaction and fluorescence-activated cell sorter, respectively. (B) Suppression of cytarabine-stimulated CD25 expression by PIM inhibitor. Cells were treated with compound C at the indicated concentrations in the presence of cytarabine (30 nM). CD25 expression was then examined by quantitative reverse-transcription polymerase chain reaction and fluorescence-activated cell sorter. (C) Fluorescence-activated cell sorter analysis of CD25 expression in the parental (i, iii) and cytarabine-resistant KG1 cell lines (ii, iii). KG1 cells were cultured in the presence of 30 nM cytarabine for 3 months. The survived cells were treated with cytarabine. The resistance was confirmed by the increased IC50 value to cytarabine compared with that of the parental line. (D) CD25 mRNA expression in the cytarabine-resistant cell line. (E) Regulation of CD25 expression in the cytarabine-resistant line. KG1 cells were treated with either STAT5A/B siRNA or control siRNA for 72 hours, and CD25 mRNA levels were analyzed by quantitative reverse-transcription polymerase chain reaction (ii). STAT5 knockdown was confirmed by western blot (i). (F) Differential response of cytarabine-resistant line to cytarabine or PIM inhibitor. KG1 parental and cytarabine-resistance cell lines were treated with serial diluted (1:3) cytarabine or compound C. The cytarabine-resistant cell line is more sensitive to PIM kinase inhibition than the parental line (ii). IC50 values are indicated.
Cytarabine-resistant cells express higher level of CD25 and are more sensitive to PIM kinase inhibition. (A) The effect of cytarabine on CD25 expression in cells. KG1 cells were treated with cytarabine at the indicated concentrations. The expression of CD25 was examined at the mRNA (i) and protein (ii) levels by quantitative reverse-transcription polymerase chain reaction and fluorescence-activated cell sorter, respectively. (B) Suppression of cytarabine-stimulated CD25 expression by PIM inhibitor. Cells were treated with compound C at the indicated concentrations in the presence of cytarabine (30 nM). CD25 expression was then examined by quantitative reverse-transcription polymerase chain reaction and fluorescence-activated cell sorter. (C) Fluorescence-activated cell sorter analysis of CD25 expression in the parental (i, iii) and cytarabine-resistant KG1 cell lines (ii, iii). KG1 cells were cultured in the presence of 30 nM cytarabine for 3 months. The survived cells were treated with cytarabine. The resistance was confirmed by the increased IC50 value to cytarabine compared with that of the parental line. (D) CD25 mRNA expression in the cytarabine-resistant cell line. (E) Regulation of CD25 expression in the cytarabine-resistant line. KG1 cells were treated with either STAT5A/B siRNA or control siRNA for 72 hours, and CD25 mRNA levels were analyzed by quantitative reverse-transcription polymerase chain reaction (ii). STAT5 knockdown was confirmed by western blot (i). (F) Differential response of cytarabine-resistant line to cytarabine or PIM inhibitor. KG1 parental and cytarabine-resistance cell lines were treated with serial diluted (1:3) cytarabine or compound C. The cytarabine-resistant cell line is more sensitive to PIM kinase inhibition than the parental line (ii). IC50 values are indicated.
MYC downregulation by PIM kinase inhibitors consistently corresponds with cellular sensitivity to PIM inhibition
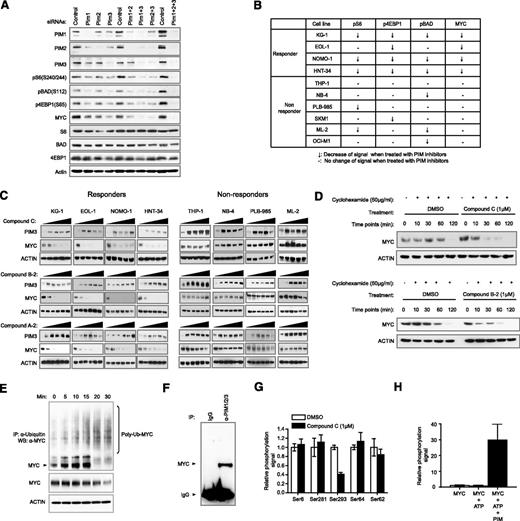
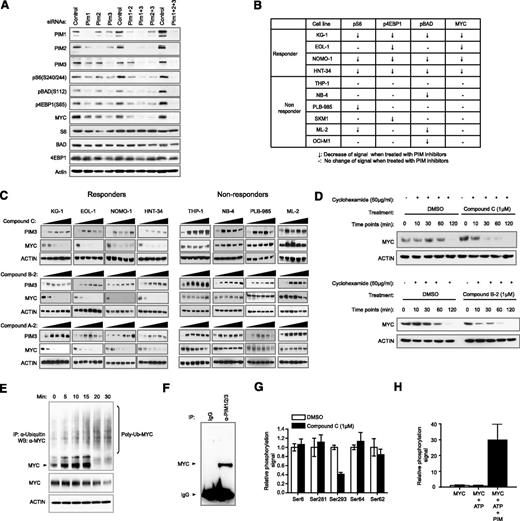
PIM kinases have been shown to coregulate crucial signaling pathways. It is reasonable to expect a number of substrates to be directly modulated by inhibition of PIM kinases. However, which substrate is the most robust efficacy marker of PIM kinase inhibition remains to be determined. To systematically approach this problem, we evaluated changes in the expression of several well-documented substrates of PIM kinases, including BAD, 4EBP1, S6, and MYC.20-23 Knockdown of all 3 PIM kinases with siRNAs resulted in significant downregulation of pS6 (S240/244), pBAD (S112), p4EBP1(S65), and MYC (Figure 5A). Similar results were also obtained using 3 PIM inhibitors of diverse scaffolds while sparing signaling nodes not known to be directly regulated by PIM kinases (supplemental Figures 9–11). Curiously, the kinetics of modulation for the aforementioned substrates varied, with MYC and p4EBP1 protein levels readily restored within 1 hour (supplemental Figure 12). These observations may affect the selection of an appropriate pharmacodynamic biomarker to establish a pharmacokinetic/pharmacodynamic/efficacy relationship in preclinical or clinical studies. These data also demonstrate that PIM kinase activity is likely required for direct regulation of the indicated markers, albeit with distinct kinetics.
Ubiquitin pathway-mediated downregulation of MYC protein level on compound treatment correlates consistently with cell growth sensitivity toward PIM inhibition. (A) Evaluation of putative PIM substrates by siRNA knockdown. KG1 cells were treated with siRNAs against PIM1, PIM2, or PIM3, either alone or as combinations. Modulations of the indicated phosphorylation events and protein abundance were assessed by western blot. (B) A summary of changes of specified intracellular markers on treatment with PIM inhibitors in a panel of responders and nonresponders. (C) MYC protein levels in responders and nonresponders on treatment of 3 diverse PIM inhibitors. (D) Half-life of endogenous MYC on treatment of PIM inhibitors. Endogenous MYC in KG1 cells has an average half-life of between 1 and 2 hours. In the presence of a PIM inhibitor, the half-life is shortened to at least less than 30 minutes (upper). Similar results were obtained using another PIM inhibitor with a different chemical structure (lower). (E) Ubiquitination of endogenous MYC in the presence of a PIM inhibitor. The position of MYC is specified with an arrow. Upward smearing streaks are indicative of poly-ubiquitinated MYC, which are intensified at the 20- and 30-minute points. Concurrent reduction of MYC protein levels is observed. (F) PIM kinases and MYC are present in the same complex. Cellular extracts were immunoprecipitated with antibodies against PIM1, PIM2, and PIM3 in radioimmunoprecipitation assay buffer. Presence of MYC was detected by western blot. (G) Differential perturbation of MYC phosphorylation sites by PIM inhibitor. Of all detectable MYC phosphorylation sites identified by quantitative phosphoproteomic mass spectrometry, S293 is the only site demonstrated to be significantly suppressed by compound C. (H) Direct phosphorylation of MYC S293 by PIM kinases. Biochemical reaction was carried out in the presence of recombinant MYC and PIM kinases. Quantitative phosphoproteomic mass spectrometry was then used to determine the level of phospho-S293.
Ubiquitin pathway-mediated downregulation of MYC protein level on compound treatment correlates consistently with cell growth sensitivity toward PIM inhibition. (A) Evaluation of putative PIM substrates by siRNA knockdown. KG1 cells were treated with siRNAs against PIM1, PIM2, or PIM3, either alone or as combinations. Modulations of the indicated phosphorylation events and protein abundance were assessed by western blot. (B) A summary of changes of specified intracellular markers on treatment with PIM inhibitors in a panel of responders and nonresponders. (C) MYC protein levels in responders and nonresponders on treatment of 3 diverse PIM inhibitors. (D) Half-life of endogenous MYC on treatment of PIM inhibitors. Endogenous MYC in KG1 cells has an average half-life of between 1 and 2 hours. In the presence of a PIM inhibitor, the half-life is shortened to at least less than 30 minutes (upper). Similar results were obtained using another PIM inhibitor with a different chemical structure (lower). (E) Ubiquitination of endogenous MYC in the presence of a PIM inhibitor. The position of MYC is specified with an arrow. Upward smearing streaks are indicative of poly-ubiquitinated MYC, which are intensified at the 20- and 30-minute points. Concurrent reduction of MYC protein levels is observed. (F) PIM kinases and MYC are present in the same complex. Cellular extracts were immunoprecipitated with antibodies against PIM1, PIM2, and PIM3 in radioimmunoprecipitation assay buffer. Presence of MYC was detected by western blot. (G) Differential perturbation of MYC phosphorylation sites by PIM inhibitor. Of all detectable MYC phosphorylation sites identified by quantitative phosphoproteomic mass spectrometry, S293 is the only site demonstrated to be significantly suppressed by compound C. (H) Direct phosphorylation of MYC S293 by PIM kinases. Biochemical reaction was carried out in the presence of recombinant MYC and PIM kinases. Quantitative phosphoproteomic mass spectrometry was then used to determine the level of phospho-S293.
We next sought to monitor dose-dependent modulation of the aforementioned markers by 3 diverse PIM inhibitors in 4 responder and 6 nonresponder cell lines to establish a correlation among PIM kinase inhibition, downstream substrate modulation, and efficacy (supplemental Figure 13). As summarized in Figure 5B, only MYC downregulation appeared to robustly differentiate responders from nonresponders of PIM kinase inhibitors with the 10 cell lines tested. Treatment with 3 diverse PIM inhibitors led to near complete suppression in MYC protein levels in responder cells (Figure 5C). To rule out differences in cellular uptake and efflux of PIM compounds between responder and nonresponder cell lines, we monitored PIM3 protein stabilization in both groups of cell lines on treatment with PIM inhibitors. Addition of PIM inhibitors produced a similar stabilizing effect on PIM3 protein in both responder and nonresponder cell lines, implying the compounds efficiently reach their target inside the cell (Figure 5C and supplemental Figure 14).
Overexpression of PIM1 or PIM2 kinase has been reported to stabilize MYC, whereas overexpression of PIM2 kinase-dead protein has been suggested to shorten MYC protein half-life.20 We used 2 structurally distinct PIM inhibitors to test their effect on MYC protein stability. We showed that MYC protein half-life was significantly shortened when PIM kinase activity was inhibited, whereas mRNA levels remained constant (Figure 5D and supplemental Figure 15). In addition, increased ubiquitination of MYC protein was observed up to 30 minutes after compound C treatment (Figure 5E). The modulation of MYC protein levels by PIM kinases through ubiquitination is likely mediated through physical interaction, as MYC could be coimmunoprecipitated with PIM1/2/3 kinase complex in high-stringency buffer (Figure 5F). Although potential MYC phosphorylation sites regulated by PIM kinases have been previously identified through biochemical and overexpression methodologies,20 we further evaluated endogenous MYC phosphorylation in the presence of a PIM inhibitor through quantitative mass spectrometry. With 70% peptide coverage, numerous phosphorylation sites were identified, but only the phosphorylation level of S293 was significantly downregulated in the presence of compound C (Figure 5G and supplemental Figure 16). We then confirmed that MYC S293 can be phosphorylated by PIM1/2/3 kinases in a more defined in vitro biochemical assay (Figure 5H). The S293 phosphorylation of MYC protein has been previously been mapped and described in cell lines derived from hematological malignancies, although the relevant functional consequences of this MYC modification are currently unknown.40 Although our results suggest that PIM kinases prolong the stability of MYC through a direct interaction, further work is required to fully explain the regulatory mechanism involving MYC S293.
Rational combination of PIM kinase inhibitor and tyrosine kinase inhibitors achieves synergistic suppression of pathway activity and cell growth in AML
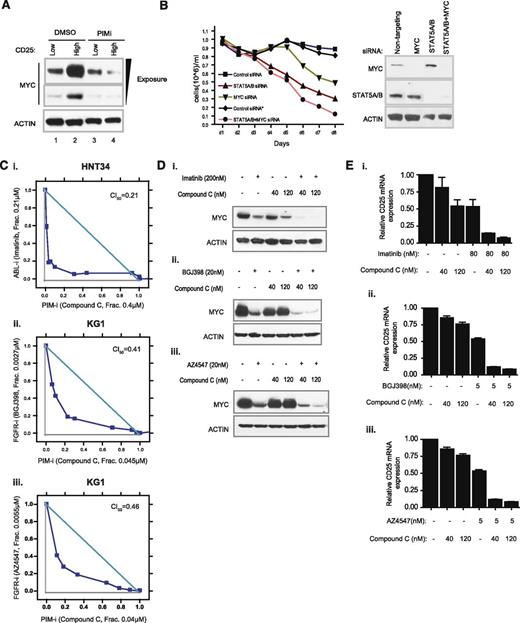
We next evaluated functional consequences of strong PIM/STAT5 and PIM/MYC physical interactions, using KG1 cells as a model. This cell line contains both CD25 high and CD25 low subpopulations, which could be efficiently separated by fluorescence activated cell sorting (supplemental Figure 17). Strikingly, MYC protein levels were significantly elevated in CD25 high KG1 cells (Figure 6A, lanes 1 and 2). Furthermore, downregulation of MYC protein by compound C was only apparent in a CD25 high subpopulation (Figure 6A, lanes 3 and 4). These results suggest that maximum STAT5 activation and MYC protein stability may be coregulated by a common mechanism that requires the activity of PIM kinases. Because both STAT5 and MYC are known to regulate cell proliferation, we aimed to test the effect of combinatorial STAT5/MYC knockdown on the growth of KG1 cells. In fact, codepletion of STAT5 and MYC by siRNAs appeared to result in further cell growth inhibition (Figure 6B).
Combination of a PIM inhibitor with tyrosine kinase inhibitors achieves synergistic effect on cancer cell growth and biomarker suppression. (A) Modulation of MYC protein expression by a PIM inhibitor in high CD25 subpopulation. KG1 cells were sorted according to the level of CD25 functional expression. The MYC protein level of the top 1% of CD25-positive cells was compared with that of CD25-negative cells in the presence of DMSO or compound C. (B) Comparison of growth suppression by depleting MYC, STAT5, or both, with siRNAs. Sustained knockdown of endogenous STAT5, MYC, or both was achieved by daily exposure of cells to Accell siRNAs (see supplemental Methods) over the course of experiments. Culture volume was diluted 1:1 every day to avoid overconfluency and to ensure the optimal growth of cells. (C) Assessment of synergistic cell growth inhibition. HNT34 (i) and KG1 (ii,iii) cell lines were treated with a combination of compound C/imatinib or compound C/pan-FGFR inhibitors. Combination index (CI) value less than 0.5 denotes significant synergism of combinatorial compound treatment. (D) Effect of compound C/imatinib or compound C/pan-FGFR inhibitor combination on MYC protein expression. (E) Effect of compound C/imatinib or compound C/pan-FGFR inhibitor combination on CD25 mRNA levels.
Combination of a PIM inhibitor with tyrosine kinase inhibitors achieves synergistic effect on cancer cell growth and biomarker suppression. (A) Modulation of MYC protein expression by a PIM inhibitor in high CD25 subpopulation. KG1 cells were sorted according to the level of CD25 functional expression. The MYC protein level of the top 1% of CD25-positive cells was compared with that of CD25-negative cells in the presence of DMSO or compound C. (B) Comparison of growth suppression by depleting MYC, STAT5, or both, with siRNAs. Sustained knockdown of endogenous STAT5, MYC, or both was achieved by daily exposure of cells to Accell siRNAs (see supplemental Methods) over the course of experiments. Culture volume was diluted 1:1 every day to avoid overconfluency and to ensure the optimal growth of cells. (C) Assessment of synergistic cell growth inhibition. HNT34 (i) and KG1 (ii,iii) cell lines were treated with a combination of compound C/imatinib or compound C/pan-FGFR inhibitors. Combination index (CI) value less than 0.5 denotes significant synergism of combinatorial compound treatment. (D) Effect of compound C/imatinib or compound C/pan-FGFR inhibitor combination on MYC protein expression. (E) Effect of compound C/imatinib or compound C/pan-FGFR inhibitor combination on CD25 mRNA levels.
Aberrantly activated tyrosine kinases are known to stimulate STAT5 activity through C-terminal tyrosine phosphorylation and to regulate MYC stability through PI3K/MAPK pathways.41 Some PIM responder cell lines identified here are known to harbor oncogenic fusion tyrosine kinases, including BCR-ABL in HNT34, FGFR1OP2-FGFR1 in KG1, and FIP1L1-PDGFRA in EOL1, which may cooperate with PIM kinases to maintain maximum STAT5 activation and prolonged MYC half-life. To evaluate whether these fusion kinases cooperate with PIM to enhance cell growth, we treated BCR-ABL-positive HNT34 cells with the combination of compound C and imatinib. Suppression of both PIM kinase and BCR-ABL activity resulted in marked synergistic inhibition of cell growth (Figure 6Ci; CI50, 0.21). A similar effect was observed when compound C was used in combination with 2 structurally distinct pan-FGFR inhibitors (NVP-BGJ398 and AZ4547) in FGFR10P2-FGFR1-positive KG1 cells (Figure 6Cii-iii; CI50, 0.41 and 0.46, respectively). Furthermore, suppression of both PIM and tyrosine kinase was more effective in reducing MYC and CD25 levels than either treatment alone (Figure 6D-E). Taken together, these observations suggest that inhibition of PIM kinase activity in the genetic context of aberrant tyrosine kinase activation could further enhance the efficacy of a targeted tyrosine kinase inhibitor.
Discussion
CD25 has been shown to be highly expressed in LSCs and positively correlated with both progenitor cell surface marker CD34 and incidence of minimal residual disease.2,5 In addition, 3 independent reports using diverse patient cohorts established a significant positive correlation between the percentage of the CD25 positive leukemic blast population (>10% or >20%) and poor overall survival or relapse-lapse free survival.3-5 Because CD25 is transcriptionally regulated by STAT5, high CD25 expression in a leukemic blast subpopulation likely reflects STAT5 activation status. Both STAT5 transcription factors and MYC oncogene have been demonstrated to play critical roles in the expansion or maintenance of hematopoietic stem cells, LSCs, or leukemia-initiating cells.25,27,28 Interestingly, our data suggest that STAT5 activity and MYC stability could be coregulated, as the cellular population expressing high CD25 also displays high levels of MYC protein (Figure 6A).
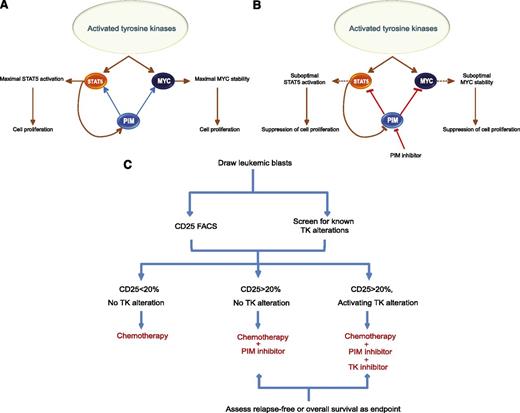
In this report, we demonstrate that CD25 is a mechanistically relevant predictive biomarker of PIM kinase inhibition. Our findings in cell lines were further supported by data obtained in patient-derived primary leukemic blast samples. Our results also show that PIM kinases are likely required to sustain the optimal activation of STAT5 and enhance MYC protein stability in AML, thereby contributing to the maintenance of cell proliferation. This observation is also supported by the synergistic regulation of STAT5 activity and MYC stability by PIM kinases and aberrantly activated tyrosine kinases. (Figure 7A-B).
Schematic of proposed mechanism of action and patient stratification strategy for PIM inhibitors. (A) PIM kinases augment the tyrosine kinase-mediated STAT5 activation and MYC stability to achieve maximal proliferative effect. Abnormally activated upstream tyrosine kinases stimulate STAT5 activity and stabilize MYC protein but require PIM kinases to achieve maximal and sustained activation of these oncoproteins. (B) In the presence of anomalous tyrosine kinase activation, addition of a PIM inhibitor diminishes the activity of STAT5 and shortens the stability of MYC. A specific and potent PIM inhibitor may suppress serine phosphorylation events on STAT5 and MYC to attenuate the tyrosine kinase-mediated activation. (C) Incorporation of CD25 and genetic assessment of tyrosine kinases as patient selection biomarkers in a clinical trial.
Schematic of proposed mechanism of action and patient stratification strategy for PIM inhibitors. (A) PIM kinases augment the tyrosine kinase-mediated STAT5 activation and MYC stability to achieve maximal proliferative effect. Abnormally activated upstream tyrosine kinases stimulate STAT5 activity and stabilize MYC protein but require PIM kinases to achieve maximal and sustained activation of these oncoproteins. (B) In the presence of anomalous tyrosine kinase activation, addition of a PIM inhibitor diminishes the activity of STAT5 and shortens the stability of MYC. A specific and potent PIM inhibitor may suppress serine phosphorylation events on STAT5 and MYC to attenuate the tyrosine kinase-mediated activation. (C) Incorporation of CD25 and genetic assessment of tyrosine kinases as patient selection biomarkers in a clinical trial.
Regulation of STAT5 activity involves tyrosine phosphorylation, serine/threonine phosphorylation, dimerization, and tetramerization. In some instances, differential phosphorylation patterns could result in the recruitment of distinct protein complexes and lead to either inhibition or augmentation of STAT5 activity.42 We showed that PIM kinases and STAT5 form a tight protein complex and that inhibition of PIM kinase activity robustly downregulates STAT5 transcriptional activity by impairing STAT5 DNA binding capacity. An earlier study suggested that PIM1 negatively regulates STAT5 activity when overexpressed in cells.43 However, this conflicting observation could be a result of the high level of exogenous PIM1 expression that leads to the artificial reconfiguration of endogenous protein complexes. These observations nevertheless place PIM kinases in complex with STAT5 and imply functional interactions between the two. Although further work is required to fully describe the molecular events leading to the observed biological outcome in the current study, the dose- and time-dependent downregulation of STAT5B phosphorylation at S731 by a PIM inhibitor may provide a possible answer. The S731 residue is nestled between the positive regulatory motifs (ie, Y699 and Y725) and negative regulatory motifs (ie, Y740 and Y743) of the STAT5 C-terminal transactivation domain.44 It was shown that inactivating mutations of Y740 and 743 (Y740/743F) led to an increase in basal pY699 levels and the corresponding STAT5B transcription activity, and that the phosphorylation of S731 was able to overcome the inhibitory pY740/743 that counteracts the activating pY699.37 Our data suggest that PIM kinases could promote the phosphorylation of S731 to maximize the maintenance of STAT5 activity.
Extensive pathway interrogation using 3 diverse PIM inhibitors revealed that MYC downregulation consistently corresponds with cellular sensitivity to PIM inhibition. Our data suggest that PIM kinases positively regulate the half-life of MYC protein by direct phosphorylation and subsequent recruitment of ubiquitin ligases. The quantitative phosphoproteomics approach led to the identification of a S293 phosphorylation site on MYC protein that appears to be targeted by PIM kinases. Although S293 phosphorylation of MYC has been previously reported,40 this is the first report implicating PIM kinases in phospho-modifying MYC at this site. MYC is also known to be phosphorylated at S62 by ERK/CDK, and this modification has been shown to prolong MYC protein stability.45 However, phosphorylation of S62 was not affected on PIM inhibitor treatment (Figure 5G). It is tempting to speculate that in certain cellular contexts, S62 and S293 could synergistically promote MYC protein stability to sustain an optimal growth advantage.
PIM1/2 have been previously reported to phosphorylate MYC at S329.20 Despite an extensive phosphoproteomics effort, we were not able to detect an endogenous MYC pS329 signal in the cell line used, even though the associated peptide was readily identified.
The data presented herein suggest that a PIM inhibitor could suppress the growth of a preexisting or chemotherapy-induced CD25 positive leukemic blast subpopulation by dampening STAT5 activity and shortening MYC protein stability. Our data further indicate that PIM kinase inhibitors could be effectively used in combination with tyrosine kinase inhibitors in CD25-positive AML cells that harbor oncogenic tyrosine kinase mutations or fusions. Therefore, our results provide a mechanistic basis for a potential clinical strategy that would employ CD25 as a predictive biomarker to determine whether to add PIM inhibitors to a chemotherapy regimen and/or tyrosine kinase inhibitor treatment, depending on the genetic status of AML (Figure 7C). The molecular interrogations presented here argue for a path into the clinic for PIM inhibitors as partners in combination therapies and pave the way for the future expansion of use in indications associated with aberrant STAT5 and MYC activation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: Z.G., A.W., and S.-M.A.H. designed research, performed research, analyzed data, and wrote the article; W.Z., and J.Z. analyzed data; M.L., Q.G., C.B., M.T., F.S., J.M., C.L., S.G., B.Z., H.C., V.P., L.S., and F.A. designed research, performed research, and analyzed data; D.H. provided a vital reagent; D.W. and J.R. wrote the article; and M.D. and C.G.-E. designed research, analyzed data, and wrote the article.
Conflict-of-interest disclosure: C.L. owns stocks in Blueprint Medicines and Sanofi as well as being member of the Scientific Advisory Boards of Covagen, Mersana Therapeutics, and ProteomicFx. The remaining authors declare no competing financial interests.
The current affiliation for C.L. is Blueprint Medicines, Cambridge, MA.
The current affiliation for M.D. is Agios Pharmaceuticals, Cambridge, MA.
The current affiliation for J.M. is Agios Pharmaceuticals, Cambridge, MA.
The current affiliation for S.G. is Blueprint Medicines, Cambridge, MA.
The current affiliation for W.Z. is Pfizer, Cambridge, MA.
The current affiliation for S.-M.A.H. is Oncology Biomarker Development, Genentech Inc., South San Francisco, CA.
Correspondence: Shih-Min A. Huang, Sanofi Oncology, 640 Memorial Dr, Cambridge, MA 02139; e-mail: huang.alex@gene.com or alexhuang001@gmail.com.
References
Author notes
Z.G. and A.W. contributed equally to this study.
C.B. and M.T. contributed equally to this study.