Key Points
Generated the first comprehensive RUNX1b-specific transcriptome and binding profile in HE.
RUNX1b induces a cell adhesion and migration program prior to the downregulation of endothelial genes and the emergence of blood cells.
Abstract
During ontogeny, the transcription factor RUNX1 governs the emergence of definitive hematopoietic cells from specialized endothelial cells called hemogenic endothelium (HE). The ultimate consequence of this endothelial-to-hematopoietic transition is the concomitant activation of the hematopoietic program and downregulation of the endothelial program. However, due to the rare and transient nature of the HE, little is known about the initial role of RUNX1 within this population. We, therefore, developed and implemented a highly sensitive DNA adenine methyltransferase identification-based methodology, including a novel data analysis pipeline, to map early RUNX1 transcriptional targets in HE cells. This novel transcription factor binding site identification protocol should be widely applicable to other low abundance cell types and factors. Integration of the RUNX1 binding profile with gene expression data revealed an unexpected early role for RUNX1 as a positive regulator of cell adhesion- and migration-associated genes within the HE. This suggests that RUNX1 orchestrates HE cell positioning and integration prior to the release of hematopoietic cells. Overall, our genome-wide analysis of the RUNX1 binding and transcriptional profile in the HE provides a novel comprehensive resource of target genes that will facilitate the precise dissection of the role of RUNX1 in early blood development.
Introduction
Over a decade ago, it was shown that hematopoietic cells can arise from a specialized endothelium known as hemogenic endothelium (HE).1,2 The generation of blood cells through this endothelial-to-hematopoietic transition (EHT) is now well established and has been visualized in real-time both in vivo, in mouse and zebrafish, and in vitro using the mouse embryonic stem (ES) cell differentiation system.3-7 The transcription factor RUNX1 (AML1) is critical for the emergence of definitive blood cells in all these models.4,7-11 The different stages of the EHT are marked by the transcriptional activity of the P1 and P2 Runx1 promoters that drive the expression of the Runx1c and Runx1b isoforms, respectively.12,13 Runx1b is expressed at low levels in HE, whereas Runx1c expression is initiated later in committed blood progenitor cells.13-15
In vitro differentiation of ES cells is a powerful tool to study early events in hematopoietic development. Upon differentiation, the HE population arises from mesodermal hemangioblast precursors and is characterized by cell surface expression of cKIT and the endothelial markers VE-cadherin (CDH5) and TIE2 (TEK).16 Initially, the HE cells form tight adherent clusters, and upon EHT, they adopt the characteristic round shape of mobile hematopoietic precursors. This is associated with a gradual loss of the endothelial markers and a concomitant gain of cell surface expression of the early hematopoietic marker CD41 (ITGA2B), and subsequently, CD45.3,4 In the absence of RUNX1, differentiation is halted at the HE stage, highlighting its absolute requirement for the EHT.4
Although the ultimate consequence of RUNX1 expression in HE is the activation of hematopoietic genes and the inactivation of endothelial genes, the direct RUNX1b transcriptional targets initiating this program remain largely unknown. Several endothelial genes including Flk1, VE-cadherin, and Tie2 are downregulated at the later stages of EHT.4,17,18 However, at this stage, RUNX1c is already expressed and it is not clear if the decrease in endothelial gene expression is directly correlated with RUNX1 binding. In this context, we have recently shown that RUNX1 can indirectly repress the endothelial program, at least in part, by inducing the expression of the transcriptional repressors Gfi1 and Gfi1b in HE.19
Identification of the transcriptional program regulated by RUNX1b in HE is key to understanding the onset of hematopoiesis and would inform the design of robust protocols for the production of hematopoietic stem cells (HSCs) from ES or induced pluripotent stem cells. Previous studies aiming to reveal the earliest RUNX1b program have been hampered by several technical limitations. Firstly, HE is a rare transient subset of endothelial cells with low endogenous RUNX1b expression.4,13-15 Secondly, the RUNX1-dependent transition to hematopoiesis takes place rapidly,11,20,21 making it difficult to distinguish between immediate effects of RUNX1 in HE cells, and later, direct or indirect effects in committed blood progenitors. These impediments are highlighted by a recent study in which only 67 RUNX1 binding events were retrieved from HE by chromatin immunoprecipitation (ChIP-seq), likely representing only a small subset of the RUNX1 binding profile.14 In this study, we overcome these caveats by coupling DNA adenine methyltransferase identification (DamID) with high-throughput sequencing to map RUNX1b binding sites in HE.22,23 This alternative to ChIP relies on the deposition of “methylation tags” around the binding sites of RUNX1 by the Escherichia coli DNA adenine methyltransferase (Dam). The stability of the methylation mark and the alleviation of the need for antibodies, make this technique ideal for binding site analysis of low-expressed DNA-binding proteins in small populations. Integration of the HE-specific RUNX1-DamID binding profile with matching transcriptome data revealed that RUNX1b binds to and upregulates the expression of genes involved in cell adhesion and migration, including components of the integrin signaling pathway. This suggests that, at this early stage of hematopoietic development, RUNX1b organizes the formation of HE clusters required for the release of blood progenitors. Overall, this study provides the first comprehensive genome-wide RUNX1b target profiling in the early HE, and demonstrates that RUNX1 acts in a stage-specific fashion by activating adhesion- and migration-associated genes prior to the emergence of hematopoietic cells and the downregulation of the endothelial program.
Methods
Mouse ES cell culture
The following mouse ES lines were used in this study: Runx1wt lines (Ainv18, idam, and Bry-GFP)20,24,25 and Runx1ko lines (Ainv18Runx1−/−, iRunx1Runx1−/−, iRunx1b::damRunx1−/−, and idamRunx1−/−).4,20 All inducible lines contain the murine proximal Runx1 isoform (RUNX1b).26 ES cells were maintained and differentiated as previously described.27 Hemangioblast-enriched populations were isolated from day 3.25 embryoid bodies (EBs) by magnetic-activated cell sorting for FLK1 (Miltenyi Biotec). HE was isolated from hemangioblast cultures by magnetic-activated cell sorting (FLK1+) or fluorescence-activated cell sorting (FACS) (cKIT+, TIE2+, and CD41−).4,21 The EHT-assay was as follows: HE-enriched cells were cultured with 2 μg/mL doxycycline (dox) or vehicle. After 72 hours, cells were analyzed by FACS for CD41. Aggregation assay was as follows: hemangioblast-enriched cells were aggregated in EB medium (0.4 ng/mL IL3, 50 ng/mL KIT ligand, and 4 ng/mL IL6) in ultra-low attachment plates (CoStar),25,27 and cultured for 13 days (1 µg/mL dox or vehicle). Images were taken on a DMI 3000B with a DFC310 FX camera (Leica).
Luciferase assays
Assays were performed using the Dual-Luciferase Reporter Assay System (Promega). GeneJuice was used for transfection (Novagen) and pgl2-7.2fms has been described previously.28 Cbfβ, Runx1b, Runx1b::dam, and dam were cloned into pCDNA3.1 (Invitrogen). The CD61 region identified by RUNX1b-DamID (16 073-18 322 bp from the gene start site) and the CD61 promoter29 were polymerase chain reaction (PCR)-amplified and cloned into pGL4 (Promega). Truncated (16 073-17 405 bp) and RUNX1 binding site deleted (16 778-16 913 bp) versions were generated by digestion and re-ligation. Luciferase was measured 48 hours posttransfection of HEK293T. iRunx1Runx1−/− cells were induced with 0.3 µg/mL dox 24 hours posttransfection and luciferase measurements followed 24 hours later.
Quantitative reverse-transcription-PCR
RNA was extracted using the RNeasy Micro Kit (Qiagen). Complementary DNA was prepared using SuperScript III First-Strand Synthesis (Invitrogen). Quantitative PCR (qPCR) was performed using the Universal ProbeLibrary System (Roche) (for primers, see supplemental Table 1 on the Blood Web site).
RNA-seq and DamID-seq
Fifty bp forward reads were generated on a SOLiD 4 System (#4448379, Applied Biosystems) for DamID-seq and a SOLiD 5500xl (#4456991, Life Technologies) for RNA-seq. Libraries were generated according to the manufacturer’s instructions. For DamID-seq: HE was isolated from idamRunx1−/− and iRunx1b::damRunx1−/− lines. For each line, 3 technical and biological replicates were processed, as previously described.23 For RNA-seq: HE was isolated from idam, idamRunx1−/−, Bry-GFP, and Ainv18Runx1−/− lines. Ribosmal RNA-depleted RNA was extracted from 2 biological replicates (Qiagen RNeasy Plus Mini Kit and Epicentre Ribo-Zero Gold Kit). For datasets, visit http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE55335.
RNA-seq analysis
Strand-specific RNA-seq reads were aligned to mouse genome mm9 with SHRiMP2 (default settings).30,31 Unique reads were aligned to Ensembl (version 66) annotated genes using Annmap (only exonic reads).32 EdgeR (false discovery rate < .05; exact test) was used to call differentially expressed genes.34,35 Common genes, with the same directional change between the Runx1ko (idamRunx1−/− and Ainv18Runx1−/−)–idam and the Runx1ko- Bry-GFP comparisons were selected for subsequent analysis.
DamID peak identification pipeline (DamID-PIP)
DamID-seq reads were aligned to mouse genome mm9 using SHRiMP2 (default settings).30,31 BayesPeak33 was applied to the DamID-seq test and control samples separately. Peaks in either of the following subsets were retained: 1) Unique test sample peaks (no overlap with control sample); and 2) Overlapping peaks with a ≥ twofold change (reads per kilobase per million [RPKM] test / RPKM control) within the top 10% of regions with the greatest difference in read count (ΔRPKM = RPKM test − RPKM control). Subsequently, Poisson model-based scan statistics were used to identify peak-enriched regions (PERs).36 A 2000-bp window was scanned along each chromosome at 400 bp increments. At each position, we calculated the number of bases within the sliding window corresponding to a peak. This value, together with the length of the window, was used to compute the value of the scan statistic. Monte Carlo sampling was used to establish an approximate distribution of the scan statistic under the null hypothesis.36 Windows with a P value <.05 were designated as peak-enriched windows. Contiguous peak-enriched windows were merged to generate larger PERs. Adjacent PERs (<300 bp apart) were merged. PERs were mapped to the Ensembl genome annotation (version 66) via Annmap.32 PERs located within 30 kb of a gene (including the gene itself) were retained. When multiple genes mapped to a single PER, the closest gene was selected, generating a PER-to-gene mapping for each test sample.
Peak allocation, motif discovery, and ontology analysis
RUNX1 peaks were allocated to the following regions: upstream (−30 kb to −2 kb from transcription start site), promoter (±2 kb from transcription start site), first intron, transcript body (excluding intron 1), and downstream (up to +30 kb of transcript end). Motif discovery and co-occurrence analysis was performed using HOMER and COPS (default settings).37,38 Gene set enrichment analysis (GSEA) software was used to perform GSEA against the GSEA database39 or custom-made lists from the Gene Ontology Consortium (supplemental Table 2) were used.40 Data were also analyzed with Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, www.ingenuity.com).
Animals
All animal work was performed under regulations set out by the Home Office Legislation under the 1986 United Kingdom Animal Scientific Procedures Act.
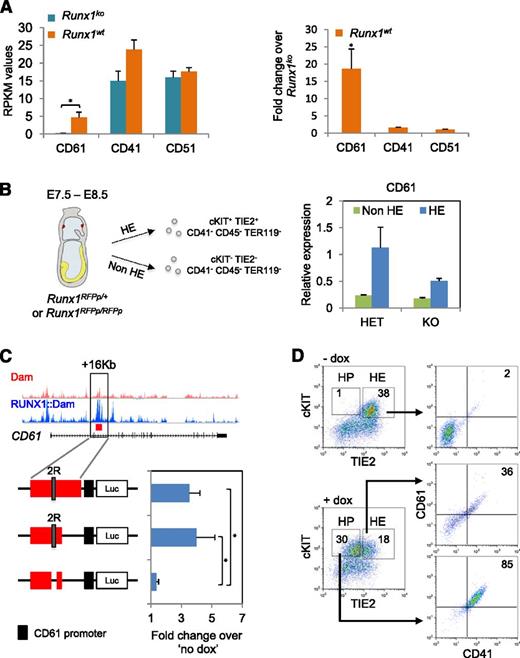
Timed matings were set up between Runx1RFPp/+ mice (red fluorescent protein [RFP] knocked-in under the proximal RUNX1 promoter, as previously described with hCD4).13 The morning of vaginal plug detection was considered day 0.5. E7.5 to E8.5 embryos were trypsinized and genotyped. HE (cKIT+, TIE2+, CD41−, CD45−, and TER119−) and non-HE (cKIT−, TIE2−, CD41−, CD45−, and TER119−) enriched cell populations were isolated by FACS.
Statistics
Unless otherwise indicated, data were evaluated using the Mann-Whitney U test and expressed as mean ± SEM. P < .05 was considered statistically significant.
Results
Establishment of RUNX1b DamID for mammalian cells
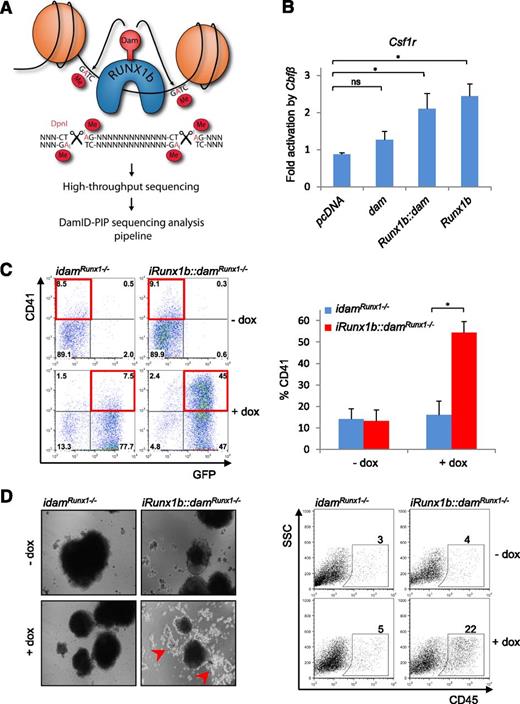
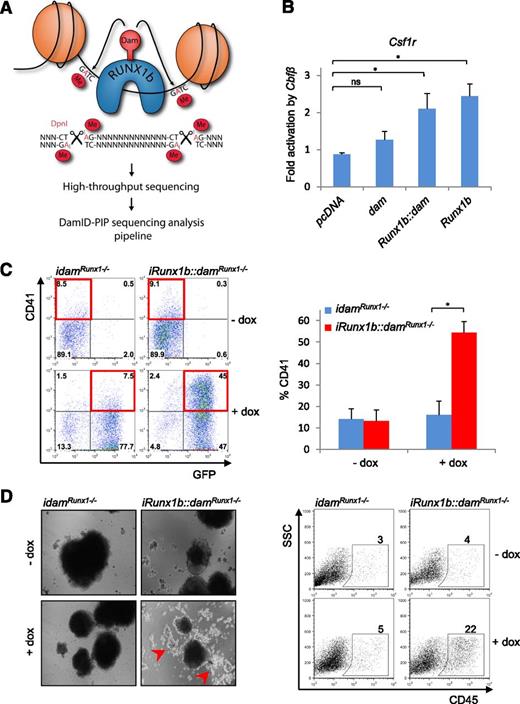
DamID is a DNA-tagging approach where a fusion between the E coli Dam and a DNA-binding protein is used to tag DNA binding sites by methylation of nearby adenines within GATC sequences22,23 (Figure 1A). We reasoned that the high processivity of Dam41 makes DamID particularly suited for profiling DNA binding of transcription factors with low endogenous expression levels in rare cell populations. We first confirmed that Dam tethering did not abrogate RUNX1 function. Optimal activity and DNA binding of RUNX1 relies on its ability to interact with CBFβ.42 Accordingly, CBFβ, combined with either RUNX1b or RUNX1b::Dam increased the transcriptional activity of the Csf1r luciferase reporter pgl2-7.2fms (a known RUNX1 target),43 demonstrating that Dam tethering does not perturb the RUNX1b-CBFβ interaction (Figure 1B). To assess if RUNX1b::Dam could drive hematopoiesis, we established 2 Runx1ko ES cell lines containing either an inducible RUNX1b::Dam-IRES-eGFP (iRunx1b::damRunx1−/−) or an inducible Dam-IRES-eGFP (idamRunx1−/−) expression cassette. Upon differentiation, Runx1ko ES cells arrest at the HE stage. In the presence of dox, the iRunx1b::damRunx1−/− line was released from this block, as illustrated by the increased frequency of CD41+ cells (Figure 1C). Furthermore, differentiation of aggregated cells under hematopoietic culture conditions promoted the release of round cells characteristic of blood formation exclusively from dox-induced iRunx1b::damRunx1−/− cells (Figure 1D). Accordingly, a high frequency of CD45+ cells was detected in induced RUNX1b::Dam cultures (Figure 1D), indicating that RUNX1b::Dam supports the generation of mature hematopoietic cells. Collectively, these experiments confirmed that RUNX1b::Dam is suitable for genome-wide RUNX1b binding profiling.
Establishment of RUNX1b-DamID for mammalian cells. (A) Schematic representation of the DamID technique. RUNX1b binding sites are marked by methylation of nearby GATC sites. Binding sites are isolated from genomic DNA using the methylation-specific restriction enzyme DpnI, amplified by PCR and subjected to high throughput sequencing. Sequencing data were analyzed by DamID-seq analysis pipeline, DamID-PIP. (B) Csf1r luciferase assay in HEK293T cells with Runx1b, Runx1b::dam, and untethered dam cotransfected with Cbfβ. *P < .05 (n = 4). (C) EHT assay in the iRunx1b::damRunx1−/− and idamRunx1−/− mouse ES lines. HE cells were cultured for 3 days in the presence or absence of dox. (Left) Representative CD41/GFP FACS plots. GFP marks Dam transgene expression. (Right) Frequency of CD41+ cells. *P < .05 (n = 4). (D) (Left) Representative images of iRunx1b::damRunx1−/− and idamRunx1−/− aggregates at day 13 of culture in the presence or absence of dox. Dox-treated iRunx1b::damRunx1−/− aggregates generate round hematopoietic cells marked by red arrows. (Right) Frequency of CD45+ cells in iRunx1b::damRunx1−/− and idamRunx1−/− aggregates at day 13 (FACS). ns, not significant.
Establishment of RUNX1b-DamID for mammalian cells. (A) Schematic representation of the DamID technique. RUNX1b binding sites are marked by methylation of nearby GATC sites. Binding sites are isolated from genomic DNA using the methylation-specific restriction enzyme DpnI, amplified by PCR and subjected to high throughput sequencing. Sequencing data were analyzed by DamID-seq analysis pipeline, DamID-PIP. (B) Csf1r luciferase assay in HEK293T cells with Runx1b, Runx1b::dam, and untethered dam cotransfected with Cbfβ. *P < .05 (n = 4). (C) EHT assay in the iRunx1b::damRunx1−/− and idamRunx1−/− mouse ES lines. HE cells were cultured for 3 days in the presence or absence of dox. (Left) Representative CD41/GFP FACS plots. GFP marks Dam transgene expression. (Right) Frequency of CD41+ cells. *P < .05 (n = 4). (D) (Left) Representative images of iRunx1b::damRunx1−/− and idamRunx1−/− aggregates at day 13 of culture in the presence or absence of dox. Dox-treated iRunx1b::damRunx1−/− aggregates generate round hematopoietic cells marked by red arrows. (Right) Frequency of CD45+ cells in iRunx1b::damRunx1−/− and idamRunx1−/− aggregates at day 13 (FACS). ns, not significant.
Genome-wide RUNX1b binding site profiling in HE
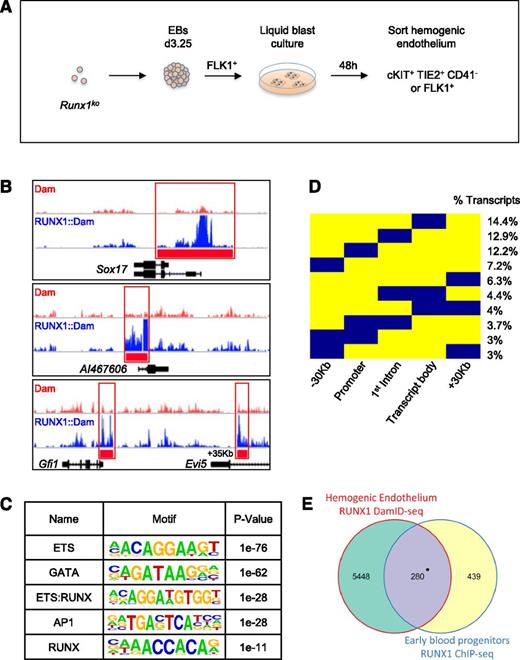
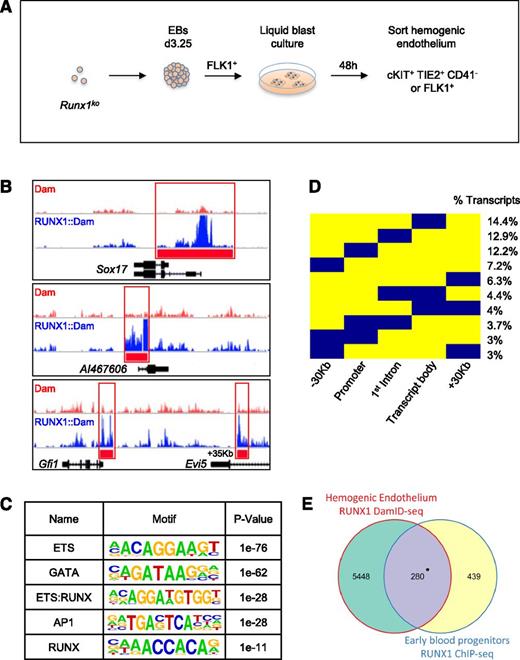
To identify early RUNX1b targets in HE, we performed DamID-seq on iRunx1b::damRunx1−/− and idamRunx1−/− HE populations (Figure 2A). All experiments were performed without inducing the DamID lines, relying only on the leakiness of the inducible system to generate trace amounts of the RUNX1b::Dam and Dam proteins. Importantly, this very low level of RUNX1b::Dam was not sufficient to rescue the Runx1ko phenotype (Figure 1C). We sequenced 3 biological replicate libraries from both iRunx1b::damRunx1−/− and idamRunx1−/− HE populations, that returned 109 and 96 million uniquely mappable reads, respectively (supplemental Figure 1A). Compared with ChIP-seq, DamID-seq generated more control peaks (untethered Dam vs ChIP IgG or input control) and wider peaks overall. Although these observations are consistent with the proximity based DNA marking underlying the DamID approach (Figure 1A),22 it also rendered existing ChIP-seq analysis packages unsuitable. We, therefore, developed a new transcription factor specific DamID analysis package (DamID-PIP). Analysis of uniquely aligned reads with DamID-PIP identified 9268 RUNX1 binding peaks that associated with 5867 genes or 7367 transcripts (supplemental Figure 1A).
Genome-wide RUNX1b binding site profiling in HE. (A) Schematic representation of the generation of HE populations for DamID-seq. Runx1ko ES cells were differentiated as EBs up to day 3.25 and sorted for FLK1 expression. FLK1+ cells were cultured in liquid blast conditions for 48 hours and then sorted either for cKIT+, TIE2+, CD41−, or FLK1+ to enrich for the HE population. (B) Raw DamID-seq read data from RUNX1b::Dam and control Dam samples in Runx1ko HE. Red bars indicate the RUNX1b::Dam peaks identified by DamID-PIP showing binding to the known RUNX1 targets Sox17, AI467606, and Gfi1. (C) Motif discovery analysis on RUNX1 DamID peaks in Runx1ko HE. Table shows the top 5 most enriched motifs. (D) Binary heat map depicting the location of RUNX1 peaks (in blue) relative to transcript coordinates. Each transcript bound by RUNX1 was divided into 5 regions: −30 kb region upstream of transcription start site (TSS), promoter (±2 kb from TSS), first intron, transcript body without the 1st intron, and +30 kb region downstream of transcript end. Numbers indicate percentage of RUNX1-bound transcripts (total number of transcripts = 7367). Only peak distribution patterns encompassing ≥ 3% of transcripts are shown. (E) Venn diagram showing overlap of RUNX1 target genes determined by RUNX1 DamID-seq in Runx1ko HE and by RUNX1 ChIP-seq in EBP (combination of HE and hematopoietic precursors). *P = 1.84−54.
Genome-wide RUNX1b binding site profiling in HE. (A) Schematic representation of the generation of HE populations for DamID-seq. Runx1ko ES cells were differentiated as EBs up to day 3.25 and sorted for FLK1 expression. FLK1+ cells were cultured in liquid blast conditions for 48 hours and then sorted either for cKIT+, TIE2+, CD41−, or FLK1+ to enrich for the HE population. (B) Raw DamID-seq read data from RUNX1b::Dam and control Dam samples in Runx1ko HE. Red bars indicate the RUNX1b::Dam peaks identified by DamID-PIP showing binding to the known RUNX1 targets Sox17, AI467606, and Gfi1. (C) Motif discovery analysis on RUNX1 DamID peaks in Runx1ko HE. Table shows the top 5 most enriched motifs. (D) Binary heat map depicting the location of RUNX1 peaks (in blue) relative to transcript coordinates. Each transcript bound by RUNX1 was divided into 5 regions: −30 kb region upstream of transcription start site (TSS), promoter (±2 kb from TSS), first intron, transcript body without the 1st intron, and +30 kb region downstream of transcript end. Numbers indicate percentage of RUNX1-bound transcripts (total number of transcripts = 7367). Only peak distribution patterns encompassing ≥ 3% of transcripts are shown. (E) Venn diagram showing overlap of RUNX1 target genes determined by RUNX1 DamID-seq in Runx1ko HE and by RUNX1 ChIP-seq in EBP (combination of HE and hematopoietic precursors). *P = 1.84−54.
The RUNX1 binding peaks contained previously identified RUNX1 binding regions, such as the promoters of AI46760644 and Sox17,14,21 and the −35 kb enhancer of Gfi1,19,45 thus providing support for the validity of our data (Figure 2B, red bars). In addition, analysis of over-represented sequence motifs (Figure 2C) identified ETS, GATA, and AP1 as the top enriched motifs. All of these factors have been shown to associate with RUNX1.14,21,46-48 RUNX1 itself was also among the top enriched motifs as a hybrid ETS-RUNX motif (Figure 2C). The RUNX motif alone was also significantly enriched but did not appear at the top of the list. This is consistent with the observation that RUNX1 can bind indirectly through interactions with other factors such as GATA2, ERG SCL, and AP1.46,48 In agreement with this, we also found by motif co-occurrence analysis, that the AP1 motif closely localized with all the top enriched motifs (supplemental Figure 1B). Subsequent analysis of the RUNX1b peak distribution relative to gene transcript coordinates revealed that 28.8% of the transcripts were exclusively bound by RUNX1b in the promoter and/or the first intron (Figure 2D). We also found that 47% of the transcripts were bound at more than 1 site (Figure 2D). This suggests that RUNX1b exerts a significant part of its regulatory function through binding to cis-regulatory elements.
We next compared the RUNX1 HE DamID peaks to binding events previously mapped by ChIP-seq in ES-derived early blood progenitors (EBP). EBPs are a mixed population of HE and early blood precursors (populations 2, 3, and 4 as described by Tanaka et al).14 A total of 39% of RUNX1 peaks previously mapped in EBP were retrieved in our DamID analyses (Figure 2E). The 280 genes bound by RUNX1 in both populations were mainly associated with regulation of gene expression and a hematopoietic signature (supplemental Figure 1C). The unique RUNX1 bound genes in EBP were not significantly enriched for any specific biological function (supplemental Figure 1C). In contrast, genes uniquely identified by HE RUNX1b DamID-seq were significantly enriched for cellular growth and proliferation, cellular movement, and cardiovascular system development, as was the DamID-seq HE gene set as a whole (supplemental Figure 1C-D).
RUNX1 expression correlates with an endothelial-associated cell adhesion and migration program in HE
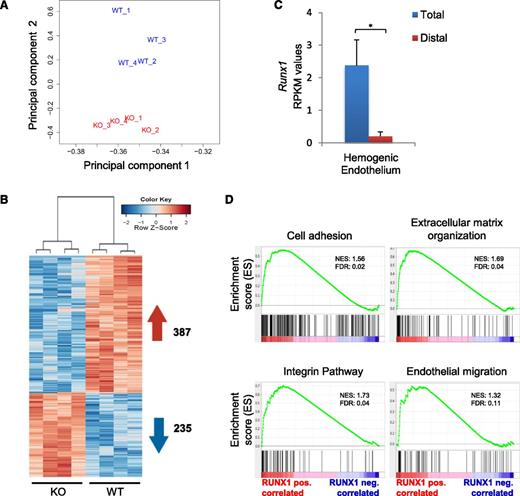
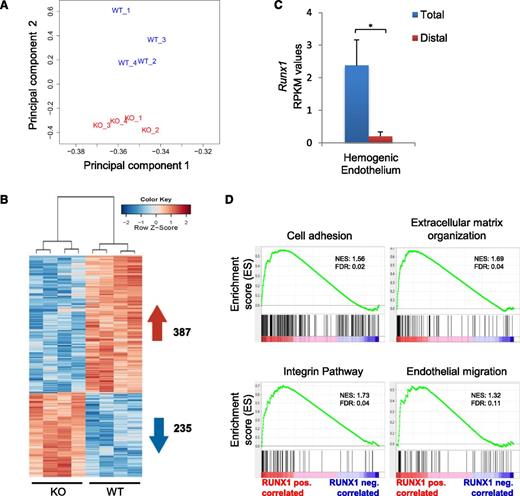
To further explore the unique binding profile of RUNX1b, we complemented our RUNX1 binding profile with transcriptional data from HE cell populations generated from Runx1wt and Runx1ko ES cells (Figure 2A). The expression patterns of Runx1wt and Runx1ko HE samples were clearly separated based on principal component analysis, and 622 genes were found to be differentially expressed (false discovery rate <.05) between the 2 populations (Figure 3A-B). Runx1c transcription was absent in our wild-type (WT) samples, confirming that we profiled the early RUNX1b-positive HE (Figure 3C). The 235 genes negatively correlated with RUNX1 did not show significant enrichment for any biological function (supplemental Figure 2A). In contrast, IPA for the 387 genes positively correlated with RUNX1 expression identified cellular movement as the most enriched function, while genes associated with hematopoietic development were only moderately enriched (supplemental Figure 2B). Likewise, GSEA showed significant correlation with cell adhesion, extracellular matrix (ECM) organization, integrin signaling, and endothelial migration datasets (Figure 3D). These findings are in agreement with the HE DamID-seq (supplemental Figure 1C-D) and the morphology of the WT and knockout (KO) HE clusters. WT HE cells assemble in tightly packed clusters, whereas Runx1 KO HE cells assemble in clusters where individual cells are easily distinguishable (supplemental Figure 3A-B and supplemental videos 1-4). Together, these data suggest an early role for RUNX1b in regulating the expression of genes promoting cell movement, cell-cell adhesion, and cell-matrix contact in HE cells prior to the HE losing its endothelial identity.
RUNX1 expression correlates with the induction of an endothelial-associated cell adhesion and migration program in the HE. (A) Principal component analysis of RNA-seq expression data from all biological replicates of Runx1wt and Runx1ko HE populations (n = 4). (B) Heat map depiction of all differentially expressed genes between Runx1wt and Runx1ko HE. A total of 387 genes (in red) are positively correlated with RUNX1 expression and 235 genes (in blue) are negatively correlated. (C) RPKM-mapped reads expression values of RUNX1 exons in Runx1wt HE. Total expression was based on exon 8 and RUNX1 distal expression on exons 1 and 2. *P < .001. (D) GSEA showing significantly enriched biological processes and signaling pathways in the gene set positively correlated with RUNX1 expression in HE.
RUNX1 expression correlates with the induction of an endothelial-associated cell adhesion and migration program in the HE. (A) Principal component analysis of RNA-seq expression data from all biological replicates of Runx1wt and Runx1ko HE populations (n = 4). (B) Heat map depiction of all differentially expressed genes between Runx1wt and Runx1ko HE. A total of 387 genes (in red) are positively correlated with RUNX1 expression and 235 genes (in blue) are negatively correlated. (C) RPKM-mapped reads expression values of RUNX1 exons in Runx1wt HE. Total expression was based on exon 8 and RUNX1 distal expression on exons 1 and 2. *P < .001. (D) GSEA showing significantly enriched biological processes and signaling pathways in the gene set positively correlated with RUNX1 expression in HE.
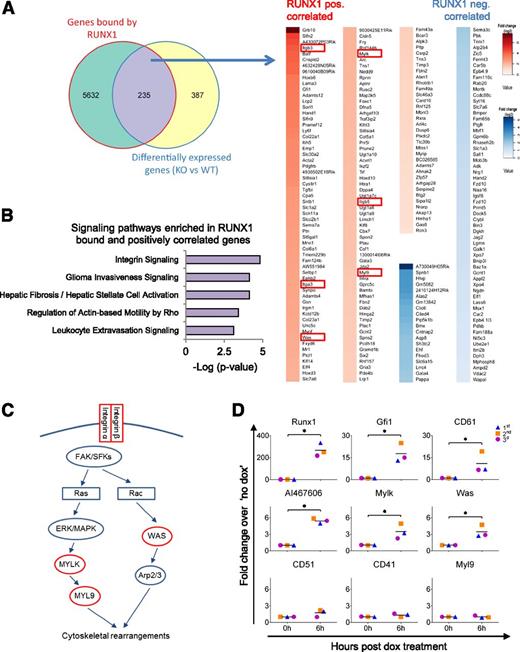
RUNX1 binds to and positively regulates genes involved in cell adhesion, cell migration, and cell-ECM interaction in HE
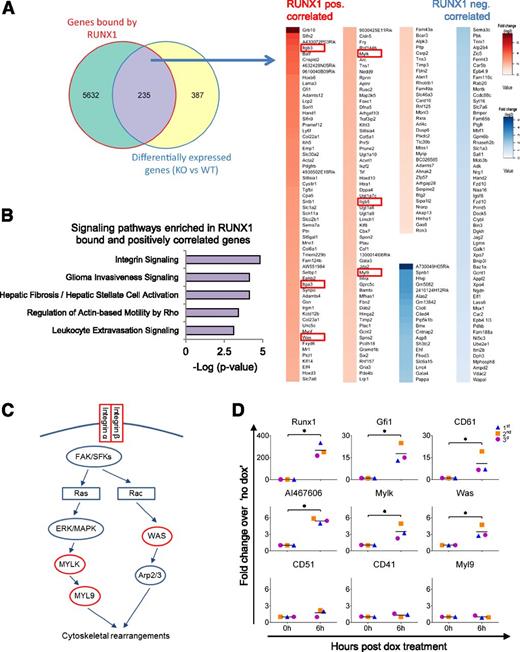
We next compared the RUNX1b DamID-seq and the RUNX1 RNA-seq datasets, and found 235 genes that were both bound and differentially expressed in the presence or absence of RUNX1 (Figure 4A and supplemental Table 3). We found 80 differentially expressed genes that negatively correlated with the presence of RUNX1. Within this subset, there was only a low level of significant enrichment for specific biological functions (supplemental Figure 4A and supplemental Table 4). In contrast, the 155 genes bound and positively correlated with RUNX1 expression were clearly associated with cell adhesion (Itga3, Itgb3, Itgb5, Lama3, Nedd9, Pcdh18, Cldn5, Synpo, Sntb1, and Tns3), cellular movement (Ephb2, Plau, Itgb3, and Bcar3), and interaction with the ECM (Adamts12, Adamts4, Adamts7, Col5a1, Col6a1, Fbln2, Lama3, Fbn2, Spon2, Timp2, Timp3, and Tgfbi) (Figure 4A, supplemental Figure 4B, and supplemental Table 5). In this dataset, we also found genes associated with vascular development (Acvrl1, Pdgfrb, Ptn, and Hand1) and regulation of transcription (Btg2, Gata3, Ikzf2, Jdp2, Klf8, Batf, Cbx7, Foxc1, Gfi1, Hand1, Hoxb6, Hoxd10, Hoxd3, Hmga2, Elf4, Rxra, and Zfp57) (Figure 4A).
RUNX1 binds to and positively regulates genes involved in cell adhesion, cell migration, and cell-ECM interaction in the HE. (A) (Left) Venn diagram showing the overlap between RUNX1-bound and differentially expressed genes in HE. (Right) Heat map depiction of the 235 RUNX1-bound and differentially expressed genes. RUNX1 positively correlated genes are marked in red shades and negatively correlated genes are marked in blue shades. The genes are ordered according to fold change in differential expression (from high to low). Red boxes show components of the integrin signaling pathway. (B) Enriched signaling pathways in RUNX1-bound positively correlated genes as determined by IPA. (C) Depiction of integrin signaling pathway with RUNX1-bound positively correlated targets shown in red. Integrins mediate signaling through the focal adhesion kinase and SRC-family kinases to mediate cytoskeletal rearrangements by either activating the Arp2/3 complex through the Wiskott–Aldrich syndrome protein, or by activating the myosin light (MYL9 or MLC) chain through phosphorylation from the MYL9 kinase (MYLK or MLCK). (D) qPCR analysis on iRunx1Runx1−/− HE cells at 0 and 6 hours postdox treatment. Graphs show fold change relative to no dox treatment of 3 biological replicates. *P < .05. Paired Student t test was used for statistical analysis.
RUNX1 binds to and positively regulates genes involved in cell adhesion, cell migration, and cell-ECM interaction in the HE. (A) (Left) Venn diagram showing the overlap between RUNX1-bound and differentially expressed genes in HE. (Right) Heat map depiction of the 235 RUNX1-bound and differentially expressed genes. RUNX1 positively correlated genes are marked in red shades and negatively correlated genes are marked in blue shades. The genes are ordered according to fold change in differential expression (from high to low). Red boxes show components of the integrin signaling pathway. (B) Enriched signaling pathways in RUNX1-bound positively correlated genes as determined by IPA. (C) Depiction of integrin signaling pathway with RUNX1-bound positively correlated targets shown in red. Integrins mediate signaling through the focal adhesion kinase and SRC-family kinases to mediate cytoskeletal rearrangements by either activating the Arp2/3 complex through the Wiskott–Aldrich syndrome protein, or by activating the myosin light (MYL9 or MLC) chain through phosphorylation from the MYL9 kinase (MYLK or MLCK). (D) qPCR analysis on iRunx1Runx1−/− HE cells at 0 and 6 hours postdox treatment. Graphs show fold change relative to no dox treatment of 3 biological replicates. *P < .05. Paired Student t test was used for statistical analysis.
Overall, integrin signaling was the top enriched canonical pathway in this gene set (Figure 4B). Integrins regulate diverse biological processes including cell migration/adhesion and signaling from the ECM,49 thus, linking the main functions that are positively regulated by RUNX1 in HE. We, therefore, decided to focus on the regulation of integrin signaling by RUNX1. Our analysis indicated that RUNX1 regulates the expression of α and β integrins (Itga3 [CD49C], Itgb3 [CD61], and Itgb5), as well as downstream targets of integrin signaling (Was, Mylk or Mlck, and Myl9 or Mlc)50,51 (Figure 4A,C). To validate that RUNX1 can directly regulate components of the integrin signaling pathway, we assessed their transcription in inducible Runx1ko HE cells (iRunx1Runx1−/−).4 A short 6 hours of dox treatment was enough to induce the expression of Runx1, and the known RUNX1 transcriptional targets Gfi119,45 and AI46760644 (Figure 4D). Similarly, expression of the integrin pathway components, CD61, and downstream targets Was and Mylk, but not Myl9, were upregulated during this short window of induction. These data validate that, in HE, the integrin signaling pathway can be regulated by RUNX1.
RUNX1 directly controls transcription of integrin β3 (CD61) in HE
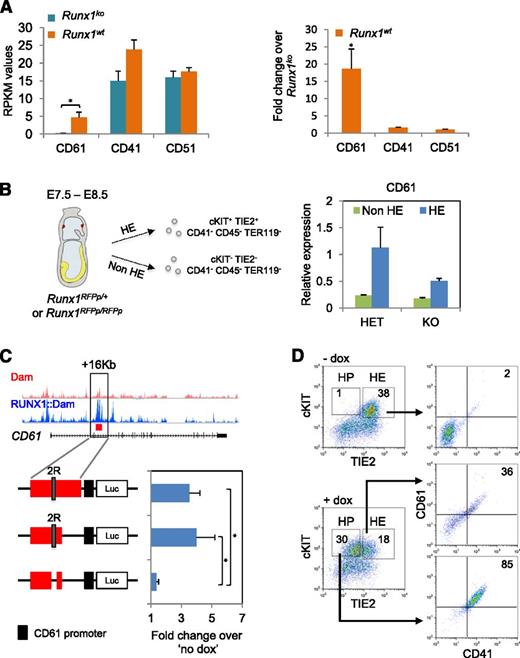
Because both the HE RNA-seq data (Figure 5A) and the Runx1 induction experiment (Figure 4D) demonstrated that CD61 was strongly upregulated by RUNX1, we focused on it in more detail. Firstly, we validated CD61 as a RUNX1 target in vivo by isolating HE- enriched populations from E7.5 to E8.5 RFP Runx1 knock-in mouse embryos13 (Figure 5B). We observed a 2× reduction of CD61 messenger RNA (mRNA) levels in Runx1ko-enriched HE cells (Runx1RFPp/RFPp) compared with their normal counterparts (Runx1RFPp/+). Similar results were obtained using a LacZ Runx1 knock-in strain9 (supplemental Figure 5A). Interrogation of the DamID-seq data identified a 2.3 kb RUNX1 bound region located 16 kb downstream of the CD61 gene start site (Figure 5C). We cloned this region together with the CD61 promoter into a luciferase reporter construct and evaluated its activity in FLK1+ cells derived from iRunx1Runx1−/− ES. Re-expression of Runx1 resulted in a 3.5-fold increase of luciferase activity (Figure 5C). Truncation of the construct revealed that the Runx1 responsive region lies within the first 1500 bp. Subsequent removal of 2 adjacent Runx1 sites completely abolished the luciferase activity, thus, demonstrating that RUNX1 can regulate transcription of CD61 through this +16 kb cis-regulatory element.
RUNX1 directly controls transcription of integrin β3 (CD61) in the HE. (A) (Left) RPKM mapped reads expression values of CD61, CD41, and CD51 genes in Runx1wt and Runx1ko HE. (Right) Fold change of RPKM values of CD61, CD41, and CD51 genes in Runx1wt HE normalized against Runx1ko. *P < .05. (B) qPCR for CD61 mRNA expression levels in non-HE (cKIT−, TIE2−, CD41−,CD45−, and TER119−) and HE (cKIT+, TIE2+, CD41−, CD45−, and TER119−) enriched populations sorted from E7.5 and E8.5 heterozygous (HET) (RUNX1RFPp/+) and KO (RUNX1RFPp/RFPp) embryos. Data are normalized against the HET HE population. (C) Luciferase assay with the full length and truncated +16 kb CD61 cis-regulatory element cloned upstream of the CD61 promoter. The “2R” depicts the 2 active RUNX1 binding motifs. The activity of all luciferase vectors was tested in iRunx1Runx1−/− HE 24 hours postdox induction. Values were normalized against “no dox” treatment. *P < .05. (D) cKIT/TIE2 and CD61/CD41 FACS plots of iRunx1Runx1−/− HE cells in the presence or absence of dox. HP, hematopoietic progenitors.
RUNX1 directly controls transcription of integrin β3 (CD61) in the HE. (A) (Left) RPKM mapped reads expression values of CD61, CD41, and CD51 genes in Runx1wt and Runx1ko HE. (Right) Fold change of RPKM values of CD61, CD41, and CD51 genes in Runx1wt HE normalized against Runx1ko. *P < .05. (B) qPCR for CD61 mRNA expression levels in non-HE (cKIT−, TIE2−, CD41−,CD45−, and TER119−) and HE (cKIT+, TIE2+, CD41−, CD45−, and TER119−) enriched populations sorted from E7.5 and E8.5 heterozygous (HET) (RUNX1RFPp/+) and KO (RUNX1RFPp/RFPp) embryos. Data are normalized against the HET HE population. (C) Luciferase assay with the full length and truncated +16 kb CD61 cis-regulatory element cloned upstream of the CD61 promoter. The “2R” depicts the 2 active RUNX1 binding motifs. The activity of all luciferase vectors was tested in iRunx1Runx1−/− HE 24 hours postdox induction. Values were normalized against “no dox” treatment. *P < .05. (D) cKIT/TIE2 and CD61/CD41 FACS plots of iRunx1Runx1−/− HE cells in the presence or absence of dox. HP, hematopoietic progenitors.
CD61 needs to heterodimerize with CD41 or CD51 to form a functional receptor.52 The CD61/CD51 heterodimer is involved in the regulation of endothelial cell migration and proliferation during neovascularization,53-55 whereas the detection of the CD61/CD41 heterodimer at the cell surface marks the first hematopoietic progenitors generated in vivo and in vitro.56-60 In contrast to CD61, both CD51 and CD41 were already present in Runx1ko HE cells and were not differentially expressed compared with WT populations (Figure 5A). Accordingly, re-expression of RUNX1 in Runx1ko HE cells did not alter the CD41 or CD51 mRNA expression levels (Figure 4D). Consistent with this observation, we detected CD41 transcription in the developing HE prior to the emergence of CD41 on the cell surface by using a CD41::Venus reporter line (supplemental Figure 5B). We also detected CD41 protein expression in Runx1ko HE cells by immunofluorescence and western blot analysis (supplemental Figure 5C and supplemental videos 1-4). Finally, we analyzed cell surface protein expression of the CD41/CD61 heterodimer, following Runx1 induction in HE-arrested Runx1ko cells (Figure 5D). Cells blocked at the HE stage did not stain for CD41 or CD61 but re-expression of RUNX1 resulted in the generation of hematopoietic precursors (HP, cKIT+, and TIE2low/−), which were double-positive for CD41/CD61 (Figure 5D). Overall, these data suggest that in HE, RUNX1 directly controls the transcription of the CD61 integrin, which is required for the presence of CD41 at the cell surface.
Discussion
The scarcity of these cells and the low level of endogenous RUNX1b expression have hampered previous studies aimed at identifying RUNX1b targets in HE. To circumvent these limitations, we developed a novel protocol combining DamID technology, high throughput sequencing, and a new analysis pipeline (ie, DamID-PIP). Compared with a ChIP-based approach,14 DamID-seq significantly increased the number of known RUNX1 binding sites in HE. This brings the number of binding events in line with those found for RUNX1 in more accessible hematopoietic cells.46,47 Our results confirm that our protocol overcomes the restrictions of ChIP-based binding site identification, and allows detection of a large number of binding events for low abundance transcription factors in small mammalian cell populations.
Integration of the HE-specific DamID-seq binding profile with matching transcriptome data revealed the absence of a strong hematopoietic signature and indicated that RUNX1 activates genes associated with cell adhesion, ECM organization, integrin signaling, and endothelial migration at this stage of differentiation. This is in line with a recent study that revealed an active endothelial developmental potential and gene signature in HE isolated from E8.5 murine embryos.15 Subsequently, from E9.5 onwards, a decline of the endothelial potential and the initiation of hematopoietic specification were observed.15 These findings suggest that we profiled early HE development and identified a novel function of RUNX1b prior to its reported role in the up and downregulation of hematopoietic and endothelial genes, respectively.
We propose that at the onset of the EHT, RUNX1b orchestrates HE-cell positioning and assembly in preparation of blood cell release. This is consistent with studies reporting perturbations in the morphology of HE cells in the absence of RUNX1. In Runx1ko mouse embryos, the endothelial cells in the floor of the dorsal aorta (a major site of EHT), have been reported to be flatter and more elongated compared with their normal counterparts.9 We observed a similar phenotype in the ES cell system with Runx1ko HE cells assembling into “loose clusters” (supplemental Figure 3A-B, supplemental videos 1-4, and supplemental videos 1 and 3 in Lancrin et al4 ).
Integrins are key players in the regulation of cell adhesion and migration.49 Accordingly, we identified integrin signaling as the most significantly upregulated canonical pathway by RUNX1b. This is exemplified by the RUNX1-dependent expression of CD61 through a +16 kb cis-regulatory element. CD61 forms a functional cell surface receptor by heterodimerizing with integrin subunits CD41 or CD51. The CD61/CD51 heterodimer is involved in neovascularization through positive regulation of endothelial cell migration and proliferation, a role that could potentially support the organization and formation of HE clusters.54 The CD61/CD41 pair marks the generation of blood precursors both in vitro and in vivo.56-60 CD41, which exclusively dimerizes with CD61, has been proposed to mediate adhesion of hematopoietic progenitors in the bone marrow niche.57 More recently, CD41 has been shown to be important for the maintenance of HSC activity in the mouse embryonic aorta.61 Interestingly, we found both CD41 and CD51 to be expressed independently of RUNX1. This suggests that in HE, RUNX1b may modulate cell surface expression of these integrins via CD61 transcription.
In conclusion, we present in this study, the first comprehensive genome-wide RUNX1b binding profile in HE and reveal a previously unidentified role for RUNX1 in activating adhesion- and migration-associated genes prior to the emergence of hematopoietic cells. This study serves as a unique resource of candidate RUNX1 target genes at the onset of hematopoiesis. Mining this dataset will not only enrich our knowledge of this crucial developmental process, but it can also inform other research fields. These include cancer biology, where accumulating evidence implicates RUNX1 in the formation and metastasis of solid tumors,62-65 and regenerative medicine, with the aim to generate blood progenitors and HSCs from ES or induced pluripotent stem cells.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the following facilities of the Cancer Research UK Manchester Institute for technical support: Advanced Imaging, Biological Resources Unit, Flow Cytometry, and the Molecular Biology Core Facility. The authors also thank Dr James Bradford for initiating the DamID-PIP bioinformatics project and Mark Wappett for further developments, Prof Bas Van Steensel (Netherlands Cancer Institute; NKI) for kindly providing the DamID plasmids, and Dr Caroline Wilkinson for critically reviewing the manuscript.
The study was supported by Cancer Research UK, the Leukaemia and Lymphoma Research Foundation, and the Biotechnology and Biological Sciences Research Council.
Authorship
Contribution: M.L. and E.M. designed and performed experiments, analyzed the data, and wrote the manuscript; Y.L. and E.M. designed the bioinformatics DamID-PIP pipeline and analyzed the data; Y.L. implemented the pipeline and performed bioinformatics analysis on the sequencing data; R.P. and M.S. designed and performed experiments; C.M. and C.B. designed and supervised the research; and V.K. and G.L. designed and supervised the research project, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Georges Lacaud, Cancer Research UK Manchester Institute, The University of Manchester, Wilmslow Rd, Manchester M20 4BX, United Kingdom; email: georges.lacaud@cruk.manchester.ac.uk.
References
Author notes
M.L. and E.M. contributed equally to this study.