Key Points
A nonsense mutation in IKBKB caused the absence of IKKβ and a lack of T- and B-cell activation through their antigen receptors.
IKKβ is not necessary for development of T or B lymphocytes but is important for their activation and for the development/function of NK cells.
Abstract
Identification of the molecular etiologies of primary immunodeficiencies has led to important insights into the development and function of the immune system. We report here the cause of combined immunodeficiency in 4 patients from 2 different consanguineous Qatari families with similar clinical and immunologic phenotypes. The patients presented at an early age with fungal, viral, and bacterial infections and hypogammaglobulinemia. Although their B- and T-cell numbers were normal, they had low regulatory T-cell and NK-cell numbers. Moreover, patients’ T cells were mostly CD45RA+-naive cells and were defective in activation after T-cell receptor stimulation. All patients contained the same homozygous nonsense mutation in IKBKB (R286X), revealed by whole-exome sequencing with undetectable IKKβ and severely decreased NEMO proteins. Mutant IKKβ(R286X) was unable to complex with IKKα/NEMO. Immortalized patient B cells displayed impaired IκBα phosphorylation and NFκB nuclear translocation. These data indicate that mutated IKBKB is the likely cause of immunodeficiency in these 4 patients.
Introduction
Mutations in genes important in T-cell or in both T- and B-cell development and function cause severe combined immunodeficiency (CID), with a majority of cases caused by mutations in IL2RG, IL7RA, ADA, JAK3, RAG1, RAG2, or DCLRE1C.1,2 In contrast to severe CID, T- and B-cell development is not as severely impaired in CID. CID has been associated with hypomorphic mutations in severe CID-causing genes as well as mutations in several other genes: ZAP70, MHC class II deficiencies, PNP, ORAI-1, STIM1, NEMO, CARD11, and MALT1.3-7 However, the etiology of many patients with CID remains unknown.
The IκB kinase (IKK) complex contains 2 structurally related catalytic subunits, IKKα and IKKβ, and a regulatory subunit, IKKγ/NEMO.8,9 Null alleles of the X-linked gene encoding NEMO (IKBKG) cause incontinentia pigmenti in heterozygous females and are lethal in hemizygous males.10 Hypomorphic alleles are compatible with life in males but cause immunodeficiency and accompanying developmental abnormalities of teeth, hair, or sweat glands in many patients.3,11,12 Hypermorphic mutations in the gene encoding IkBα cause autosomal-dominant ectodermal dysplasia with T-cell immunodeficiency, undetectable memory T cells, and absence of response to CD3-T-cell receptor activation.13 The IKKα/β/NEMO complex, activated by antigen and other receptors, phosphorylates the IκB molecules, leading to subsequent NFκB nuclear translocation to activate transcription of genes involved in immune responses.14 We report a homozygous nonsense mutation in IKBKB in 4 patients with CID from 2 unrelated families and compare and contrast our findings with those in 2 other recent reports of mutations in this gene.15,16
Patient, materials, and methods
Abbreviated information is presented here. Details are provided in the supplemental Data, available on the Blood Web site.
Patients
All studies were performed with the approval of the Duke University Medical Center Institutional Review Board, and written informed consent of the patients’ parents was collected in accordance with the Declaration of Helsinki. The patients were members of 2 unrelated consanguineous families. Select clinical features are presented in Table 1, with further details available in the supplemental Data.
Immunologic phenotype analysis
Flow cytometry of peripheral blood leukocytes was performed with labeled antibodies. Lymphocyte proliferation was assessed as previously described.17
Exome sequencing, alignment, and variant calling
Exome sequencing was performed in the Genomic Analysis Facility in the Center for Human Genome Variation at Duke University. Sequencing libraries were prepared from DNA extracted from patients’ leukocytes using the Illumina TruSeq library preparation kit, following the manufacturer’s protocol.
Establishment of Epstein-Barr virus cell lines
Epstein-Barr virus (EBV)-transformed B-cell lines were established from peripheral blood leukocytes, as previously described.18
Cloning of full-length and mutant HuIKKβ, transfection, and retroviral transduction
Wild-type full-length (FL-hIKKβ) or R286X mutant IKKβ cDNA, amplified from a wild-type human IKKβ template (Addgene) with a forward primer 5′-GTGAACCGTCAGAATTGATCT-3′ and a reverse primer 5′-gagtGtttaaacACATCATGAGGCCTGCTCCA-3′ or 5′-CggaattcTCAGGGGTGCCACATCAGCATCA-3′, was cloned into the MIGR1 retroviral vector. Retroviral vectors were transfected into the Phoenix-Ampho cells to generate amphotropic retroviruses.
Cell stimulation, isolation of nuclear and cytoplasmic fractions, and immunoblot analysis
EBV-transformed B cells, which were rested in Dulbecco's phosphate buffered saline at 37°C for 30 minutes, were stimulated with phorbol myristate acetate (PMA) (20 ng/mL) or anti-human CD40 (10 μg/mL, Biolegend) at 37°C for 10 or 20 minutes. Nuclear extracts and cytosolic or total cell lysates were subjected to standard immunoblotting analysis.
Real-time quantitative polymerase chain reaction
Total RNA from peripheral blood mononuclear cells and EBV-transformed B cells was used for real-time quantitative polymerase chain reaction analysis.
Statistical analysis
Two-tail Student t test was performed (*P < .05; **P < .01; ***P < .001; Figure 1).
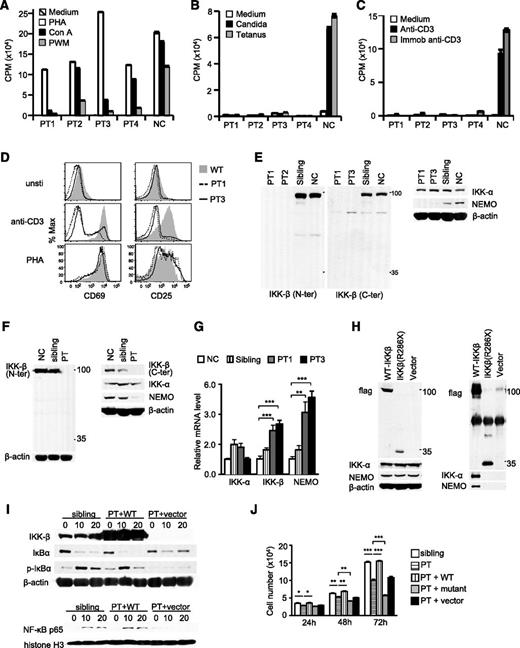
Contribution of IKKβ(R286X) mutation to CID. (A-C) Patients’ blood lymphocytes responded normally when stimulated with phytohemagglutinin (PHA) but had variably low responses to concanavalin A (Con A) and pokeweed mitogen (A). However, they failed to respond when stimulated with candida and tetanus antigens (B) or soluble or immobilized anti-CD3 (C). (D) Impaired upregulation of T-cell activation markers in patients’ CD4 T cells after overnight stimulation with plate-bound anti-CD3 or phytohemagglutinin. (E-F) Neither full-length nor truncated mutant IKKβ(R286X) protein is detectable in patients (PT), siblings, and normal peripheral blood mononuclear cells (E) and EBV-transformed B cells (F) by immunoblotting analysis with anti-N- and anti-C-terminal IKKβ antibodies. (G) mRNA levels of IKKα, IKKβ, and NEMO in PMCs detected by real-time quantitative polymerase chain reaction. (H) IKKβ(R286X) is not able to form a complex with IKKα/NEMO. Cell lysates from Phoenix-Eco cells transfected with Flag-tagged-FL-IKKβ, Flag-tagged-IKKβ(R286X), or vector control were subjected to immunoblotting analysis directly (left) or after immunoprecipitation with anti-Flag antibody conjugated agarose beads (right). (I) Defective IκBα/NFκB signaling in patient-derived B cells can be reverted by full-length (long) but not mutant IKKβ(R286X). EBV-transformed B cells of a sibling and patients stably infected with retrovirus expressing WT IKKβ or with control vector were rested in phosphate-buffered saline at 37°C for 30 minutes, followed by PMA stimulation for 10 and 20 minutes. Immunoblotting analysis of cytosolic fractions (top) or nuclear extracts (bottom) with indicated antibodies. (J) Decreased expansion of patient-derived B cells can be corrected by full-length but not mutant IKKβ. *P < .05; **P < .01; ***P < .001 determined by Student t test.
Contribution of IKKβ(R286X) mutation to CID. (A-C) Patients’ blood lymphocytes responded normally when stimulated with phytohemagglutinin (PHA) but had variably low responses to concanavalin A (Con A) and pokeweed mitogen (A). However, they failed to respond when stimulated with candida and tetanus antigens (B) or soluble or immobilized anti-CD3 (C). (D) Impaired upregulation of T-cell activation markers in patients’ CD4 T cells after overnight stimulation with plate-bound anti-CD3 or phytohemagglutinin. (E-F) Neither full-length nor truncated mutant IKKβ(R286X) protein is detectable in patients (PT), siblings, and normal peripheral blood mononuclear cells (E) and EBV-transformed B cells (F) by immunoblotting analysis with anti-N- and anti-C-terminal IKKβ antibodies. (G) mRNA levels of IKKα, IKKβ, and NEMO in PMCs detected by real-time quantitative polymerase chain reaction. (H) IKKβ(R286X) is not able to form a complex with IKKα/NEMO. Cell lysates from Phoenix-Eco cells transfected with Flag-tagged-FL-IKKβ, Flag-tagged-IKKβ(R286X), or vector control were subjected to immunoblotting analysis directly (left) or after immunoprecipitation with anti-Flag antibody conjugated agarose beads (right). (I) Defective IκBα/NFκB signaling in patient-derived B cells can be reverted by full-length (long) but not mutant IKKβ(R286X). EBV-transformed B cells of a sibling and patients stably infected with retrovirus expressing WT IKKβ or with control vector were rested in phosphate-buffered saline at 37°C for 30 minutes, followed by PMA stimulation for 10 and 20 minutes. Immunoblotting analysis of cytosolic fractions (top) or nuclear extracts (bottom) with indicated antibodies. (J) Decreased expansion of patient-derived B cells can be corrected by full-length but not mutant IKKβ. *P < .05; **P < .01; ***P < .001 determined by Student t test.
Results and discussion
All 4 infants were hypogammaglobulinemic (Table 1). Three demonstrated normal or elevated numbers of T cells and normal numbers of B cells but low numbers of switched memory B cells and NK cells. Most T cells were CD45RA+. There were abnormally low numbers of CD45RO+ T cells and CD4+CD25bright or CD4+FOXP3+ T-regulatory cells (Treg) (supplemental Figure 1).
All patient T cells displayed normal responses to phytohemagglutinin, but low responses to pokeweed mitogen or concanavalin A were seen in 3 of them (Figure 1A). Strikingly, all patients’ T cells failed to respond to candida or tetanus toxoid antigens or anti-CD3 stimulation (Figure 1B-C). Moreover, patients’ CD4 T cells were impaired for T-cell receptor-induced CD25 and CD69 upregulation (Figure 1D). NK-cell function, tested in 1 patient from each family, was impaired in both patients (data not shown).
Whole exome sequencing on patients 1 and 2 representing the 2 unrelated families identified a single candidate nonsense homozygous variant in IKBKB (R286X). The other 2 patients and the parents carried the same homozygous and heterozygous variant, respectively (supplemental Figure 1C-D), which was absent in the 998 sequenced controls and in the exome variant server public database (Exome Variant Server, National Heart, Lung and Blood Institute [NHLBI] Grand Oppportunity [GO] Exome Sequencing Project, Seattle, WA [URL: http://evs.gs.washington.edu/EVS/]).
Contrary to in healthy controls and a heterozygous sibling, anti-N- or -C-terminal IKKβ antibodies failed to detect any full-length or truncated IKKβ in patients’ peripheral blood mononuclear cells and EBV-transformed B-cell lines (Figure 1E-F) that was not caused by possible low quality of the antibody (supplemental Figure 1E). Interestingly, NEMO was virtually undetectable, but IKKα was not obviously altered in patient samples (Figure 1E-F). Both IκKβ and NEMO mRNA levels were considerably increased in patients’ peripheral blood mononuclear cells (Figure 1G). Contrary to wild-type IKKβ, IKKβ(R286X) could not associate with either IKKα or NEMO when overexpressed in Phoenix-eco cells (Figure 1H).
Patients’ EBV-transformed B cells displayed reduced IκBα phosphorylation and degradation and impaired nuclear accumulation of NFκB after stimulation with PMA, which activates the PKC-IKK-NFκB pathway (Figure 1I).19 Such defects were corrected after reconstitution with full-length but not IKKβ(R286X), suggesting that IKKβ(R286X) caused defective IκBα/NFκB activation in patient-derived B cells (Figure 1J and supplemental Figure 1F). Concordantly, expansion of patient’s EBV-transformed B cells, which was slower in vitro than their sibling’s or parents’ counterparts, could be accelerated to wild-type levels when reconstituted with full-length, but not mutant, IKKβ (Figure 1J). Interestingly, patient B cells expressing IKKβ(R286X) expanded slower than cells expressing green fluorescence protein alone, suggesting that IKKβ(R286X) exerted a dominant-negative function. At this time, it is unclear whether IKKβ(R286X) may interfere with the residual IKKα/NFκB signaling in these cells.
Together, these observations indicate that IKKβ protein is absent in the patients and that the C terminus of IKKβ mediates its association with IKKα/NEMO, which may be important for the stability of itself and of NEMO, supporting that the nonsense mutation in IKBKB caused CID in these patients. Our results indicate that IKKβ is not required for the development of T and B cells and show a novel cause of CID related to defects in the NFκB pathway. These findings highlight the diversity of phenotypes associated with mutations in different components of the NFκB pathway. For example, most patients with mutations in IKBKG had ectodermal dysplasia,20 which was absent in our IKKβ(R286X) patients even though they have almost undetectable NEMO. Interestingly, while this manuscript was in preparation/review, 2 other IKBKB mutations in CID patients were reported: one a duplication of exon 13 (IKBKB13Dup),15 and the other Y107X nonsense mutation of IKBKB.16 The clinical features of our patients were similar to those in the other 2 reports, with early infections with candida, gram-negative bacteria, viruses, and mycobacteria. The immunological features were also similar: They all have elevated numbers of naive T cells that were poorly activated by antigens and anti-CD3, low numbers of B cells that were also naive, and low numbers and function of NK cells. There was a paucity of CD45RO-positive T cells, Tregs, and γ/δ T cells. Similar to our study, mutant IKKβ protein was undetectable and NEMO was decreased in IKBKB13Dup patients.15 However, IKKα was not obviously decreased in our IKKβ(R286X) patients but was severely decreased in the IKBKB13Dup patients.15 The effect of IKKβ(Y107X) mutation on IKKα and NEMO expression was not reported.16
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Cancer Center flow cytometry core facility at Duke University for cell sorting. We acknowledge the following individuals for the contributions of control samples: Dr Gianpiero Cavalleri, Dr Norman Delanty, Dr Chantal Depondt, Dr Sanjay Sisodiya, Dr William B. Gallentine, Dr Erin L. Heinzen, Dr Aatif M. Husain, Kristen N. Linney, Dr Rodney A. Radtke, Dr Saurabh R. Sinha, Nicole M. Walley, Dr Julie Hoover-Fong, Dr Nara L. Sobreira, Dr David Valle, Dr William L. Lowe, Dr Scott M. Palmer, Dr Zvi Farfel, Dr Doron Lancet, Dr Elon Pras, Arthur Holden, Dr Elijah Behr, Dr Annapurna Poduri, Dr Patricia Lugar, Dr Rasheed Gbadegesin, Dr Michelle Winn, Dr Robert Brown, Dr Gianpiero Cavalleri, Dr Norman Delanty, Dr Chantal Depondt, Dr Yong-Hui Jiang, Dr Vandana Shashi, Kelly Schoch, Dr Eli J. Holtzman, Dr Sarah Kerns, Dr Harriet Oster, Dr Doug Marchuk, Dr Demetre Daskalakis, Dr Nicole Calakos, Dr Francis J. McMahon, Nirmala Akula, and Dr M. Chiara Manzini.
The authors would like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies which produced and provided exome variant calls for comparison: the Lung GO Sequencing Project (HL-102923), the Women's Health Initiative Sequencing Project (HL-102924), the Broad GO Sequencing Project (HL-102925), the Seattle GO Sequencing Project (HL-102926) and the Heart GO Sequencing Project (HL-103010); the National Institute of Allergy and Infectious Diseases (AI076357, AI079088, and AI101206 to X.-P.Z.); and an award from Clinical Immunology Society/Grifols (CIS/Grifols Senior Fellowship Award to T.M.). The collection of control samples and the production of sequence data were funded in part by the Epilepsy Phenome/Genome Project U01NS053998; Epi4K Project 1 – Epileptic Encephalopathies U01NS077364; Epi4K Sequencing, Biostatistics and Bioinformatics Core U01NS077303; National Institute of Allergy and Infectious Diseases Grant UO1AIO67854 (Center for HIV/AIDS Vaccine Immunology), and an award from Biogen Idec.
Authorship
Contribution: T.M. designed the study, analyzed sequencing data, and wrote the paper; J.Y. designed and performed functional studies and contributed to manuscript preparation; T.J.U. analyzed the sequencing data; H.W. participated in functional studies; M.A. referred the patients and conducted immunologic investigations; R.E.P. performed extensive immunologic studies and contributed to manuscript preparation; J.L.R. contributed to data analysis and manuscript composition; D.B.G. oversaw the whole-genome sequencing and data analysis and contributed to the manuscript; R.H.B. designed the study, wrote the paper, and performed extensive immunologic evaluations; X.-P.Z. designed functional studies, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rebecca H. Buckley, Box 2898 or 362 Jones Building, Duke University Medical Center, Durham, NC 27710, e-mail: buckl003@mc.duke.edu.
References
Author notes
T.M. and J.Y. contributed equally to this study.