Abstract
Rationale
Multiple Myeloma (MM) is characterized by the presence of multiple disease foci disseminated throughout the skeleton suggesting continuous circulation and metastasis of myeloma cells from one site of the bone marrow (BM) to another leading disease progression. However the metastatic process in MM has not been well characterized. In addition, the role of specific subclones that have the propensity for metastasis and tumor colonization in the BM niche has not been investigated. In this study, we developed a new BM metastasis xenograft model to examine clonal heterogeneity in tumor colonization of distant bone niches. We identified a set of genes that characterize potential driver genes for metastasis in MM by genomic and transcriptomic profiles of metastatic and primary tumor clones.
Methods
The model was developed by performing bilateral femur transplantation from donor SCID-bg mice to the dorsum of recipient mice of the same background. To study metastasis, the donor femurs were injected with MM cells (human MM1S, IM-9 and murine 5TGM1) and then implanted in the recipient mice. At the time of hind limb paralysis, the BM cells were flushed from the host or implanted femurs and analyzed by flow-cytometry. To investigate clonal heterogenity, IM-9 cells were transformed with four fluorescent proteins (FPs) simultaneously and sorted into fifteen subpopulations of all combinations of FPs. A mixture composed by equal proportion of all 15 FPs-labeled cells (rainbow mixture) was prepared and then used for in vivo experiment. At the time of sacrifice, clonal distribution of metastasized tumors were analyzed and the predominant clones (winner clones) were flow-sorted for genomic and transcriptomic studies. Library preparation and sequencing were performed according to manufacturer's protocols. Sequencing data was processed by bcbio_nextgen. Briefly for RNA-seq data, raw reads were aligned to reference human genome GRCh37, and gene-level read counts were calculated. Data normalization and differential expression were analyzed with DESeq2. For DNA-seq data, raw reads were aligned to GRCh37. Somatic single nucleotide variants and INDEL were called by MuTect and Indelocator, respectively.
Results
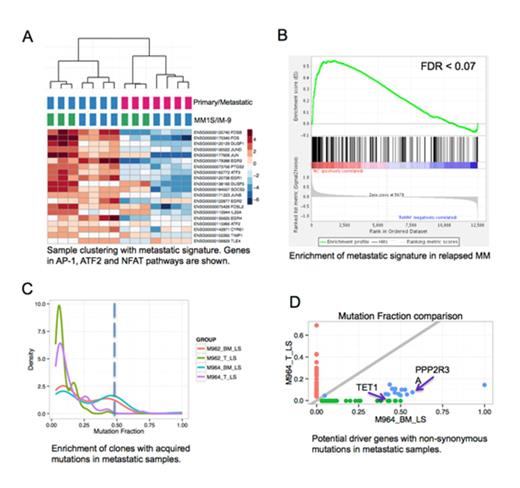
All myeloma cell lines studied were able to metastasize from the BM of transplanted femurs to the host BM and mice eventually developed paralysis after 6 to 11 weeks. Experiments using rainbow cells consistently showed that only a sub-clone of single color was able to invade and populate the host BM after metastasis, while all 15 color populations were developed in primary tumors. Interestingly, metastatic clones from different mice had similar expression profiles, although these were labeled by different colors. The studies were confirmed in a second MM cell line (MM1S) showing a similar metastasis gene signature (Fig. A). Differential expression analysis identified 238 genes significantly down regulated in both IM-9 and MM1S metastatic tumors compared to matched primary tumors (FDR < 1%). Pathway enrichment analysis indicated that AP-1, ATF2 and NFAT pathways were significantly over-represented (FDR < 5%) (Fig. A). Moreover, this metastatic signature was significantly repressed in relapsed MM patient samples compared to normal controls (FDR < 7%) using GSE6477 dataset (Fig. B). We also compared mutation fraction (MF) distributions in primary and metastatic tumors using DNA-seq data. There was only one peak in each primary tumor (MF around 0.1), while were two peaks for metastatic samples (MF at 0.1 and 0.4), strongly suggests that metastatic clones are derived from a single subclone (Fig. C). Similar results were observed with analysis of only the non-synonymous mutations (Fig. D). Out of all genes with non-synonymous mutations, we found 11 genes that are also functionally related to the metastatic signature using co-expression networks-based prioritization method. Two genes TET1 and PPP2R3A are indicated as examples (Fig. D).
Conclusions
Here we show a new model of bone metastasis that can be used to examine mechanisms of cell dissemination and colonization of the BM niche. Our studies demonstrate that specific winner subclones have a higher metastatic potential and are likely driver clones for tumor metastasis in MM. On the molecular level, a metastatic gene signature was found to be consistently down regulated in metastatic tumor samples, and 11 genes were identified as potential drivers.
Munshi:Janssen Research & Development: Membership on an entity's Board of Directors or advisory committees. Anderson:Celgene: Membership on an entity's Board of Directors or advisory committees; Millennium: Membership on an entity's Board of Directors or advisory committees; BMS: Membership on an entity's Board of Directors or advisory committees; Onyx: Membership on an entity's Board of Directors or advisory committees; Acetylon: Scientific Founder Other; Oncopep: Scientific Founder Other. Scadden:Fate Therapeutics: Consultancy, Equity Ownership. Ghobrial:Sanofi: Research Funding; Noxxon: Research Funding; BMS: Advisory board, Advisory board Other, Research Funding; Onyx: Advisory board Other.
Author notes
Asterisk with author names denotes non-ASH members.