Key Points
In differentiating ESCs, embryonic myelopoiesis is restricted by BMP signaling, independent of SMAD1/5 activity.
Blocking BMP signaling enhances Notch activity and deactivates p38MAPK, causing increased C/EBP levels and increased myeloid progenitors.
Abstract
Bone morphogenetic protein (BMP) signaling regulates early hematopoietic development, proceeding from mesoderm patterning through the progressive commitment and differentiation of progenitor cells. The BMP pathway signals largely through receptor-mediated activation of Mothers Against Decapentaplegic homolog (SMAD) proteins, although alternate pathways are modulated through various components of mitogen-activated protein kinase (MAPK) signaling. Using a conditional, short hairpin RNA (shRNA)-based knockdown system in the context of differentiating embryonic stem cells (ESCs), we demonstrated previously that Smad1 promotes hemangioblast specification, but then subsequently restricts primitive progenitor potential. Here we show that co-knockdown of Smad5 restores normal progenitor potential of Smad1-depleted cells, suggesting opposing functions for Smad1 and Smad5. This balance was confirmed by cotargeting Smad1/5 with a specific chemical antagonist, LDN193189 (LDN). However, we discovered that LDN treatment after hemangioblast commitment enhanced primitive myeloid potential. Moreover, inhibition with LDN (but not SMAD depletion) increased expression of Delta-like ligands Dll1 and Dll3 and NOTCH activity; abrogation of NOTCH activity restored LDN-enhanced myeloid potential back to normal, corresponding with expression levels of the myeloid master regulator, C/EBPα. LDN but not SMAD activity was also associated with activation of the p38MAPK pathway, and blocking this pathway was sufficient to enhance myelopoiesis. Therefore, NOTCH and p38MAPK pathways balance primitive myeloid progenitor output downstream of the BMP pathway.
Introduction
The bone morphogenetic protein (BMP) signaling pathway was identified for its role in osteogenesis,1 but has since been recognized as a key regulator of early mesoderm patterning, required for establishment of precursor populations that constitute embryonic, fetal, and adult hematopoietic lineages. Hence, the BMP ligands BMP2 and BMP4, in addition to the effector molecules SMAD1 and SMAD5, are required for normal mesodermal and hematopoietic development, and their absence causes early embryonic lethality.2-6 BMP signaling has pleiotropic effects on primitive and definitive hematopoiesis that are time- and context-dependent,7,8 which have been harnessed as tools to control developmental output in embryonic stem cell (ESC) culture-based assays.9-11 The ESC system can recapitulate, through embryoid body (EB) formation, many events of early development, including specification and proliferation of multipotent and lineage-restricted hematopoietic progenitors. Fuller understanding of BMP signaling, and how it integrates with other pathways in lineage commitment, should facilitate efforts to control stem and progenitor cell capacity for disease modeling and regenerative therapies.
BMP signaling constitutes one arm of the transforming growth factor-β (TGF-β) superfamily of signaling pathways, and propagates its biological effects largely through pathway-restricted, receptor-activated SMADs (R-SMADs). For TGF-β signaling, these are SMADs 2 and 3; for BMP, they are SMADs 1, 5, and 9 (SMAD9 was formerly called SMAD8 or MADH6). R-SMADs converge onto a limiting pool of the coactivator molecule SMAD4, which in association with a multimeric complex facilitates translocation into the nucleus and activation of target genes.12,13 In addition to R-SMAD–dependent outcomes, both TGF-β and BMP signaling have multiple effects that bypass SMAD signaling and typically proceed through 1 or more mitogen-activated protein kinase (MAPK) signaling pathways, indirectly bringing about changes in target gene transcription.14,15 For instance, TAK1 is activated by the BMP pathway to initiate a MAPK signaling cascade through p38 that regulates dorsoventral patterning in Xenopus embryos.16 BMP2 activates both p38MAPK and c-Jun N-terminal kinase (JNK) in osteoblast cultures, promoting differential regulation of alkaline phosphatase and osteocalcin production.17 Further regulation of JNK signaling by BMP ligands has been shown in models of osteogenesis18 and myocardial infarction.19 Signaling through the extracellular signal-regulated kinase 1/2 (ERK1/2) pathway is regulated by BMP in SMAD4-deficient colon cancer cells,20 as well as in osteoblast differentiation21 and endothelial sprouting.22 Finally, phosphatidylinositol 3-kinase (PI3K)/AKT signaling can act as a molecular bridge in cross-talk between BMP and WNT pathways in models of stem cell self-renewal.23
Extensive molecular cross-talk between BMP, WNT, and NOTCH signaling pathways has been described in different biological contexts, with all 3 cooperating to control specification of early ventral mesoderm derivatives that give rise to bipotential hematovascular precursors.24 These pathways further interact in precursor cells to regulate segregation of smooth muscle, blood cell, and endothelial cell progenitors.25 In ESC/EB models of yolk sac (YS) hematopoiesis, the potential to form erythroid and/or myeloid populations can be distinguished by assaying progenitor colony formation under permissive culture conditions; NOTCH signaling through Delta ligands, including specifically Dll4, has been implicated in regulating hemangioblast and endothelial development in EBs.26 Specification to the myeloid lineage is controlled by the master regulators Pu.1 and C/EBPα.27-29 Myeloid potential is further governed in part by MAPK-dependent regulation of C/EBPα activity,30 and the zebrafish Pu.1 ortholog Spi-1 marks sites of myelopoiesis that are refractive to ectopic BMP4.31
We previously used ESC/EB model systems for conditional expression or depletion of Smad1, and established its dual role in first expanding mesodermal derivatives including hemangioblasts, and then subsequently restricting hematopoietic potential.32,33 Here we report that SMAD5 contributes to SMAD1 activity, and investigate SMAD-independent control of hematopoietic potential, identifying a previously unrecognized BMP signaling axis that is propagated by activation of p38MAPK and restriction of NOTCH signaling to balance output of primitive myeloid progenitors.
Materials and methods
ESC and EB culture
ESCs were maintained on irradiated mouse embryonic fibroblast (MEF) feeder cells in Dulbecco modified Eagle medium supplemented with 20% heat-inactivated ES-qualified fetal calf serum (FCS; Hyclone), 50 IU of penicillin (Cellgro), 50 IU of streptomycin (Cellgro), leukemia inhibitory factor (LIF) (2% conditioned medium), and 1.5 × 10−4M monothioglycerol (MTG; Sigma). EBs were cultured in Iscove modified Dulbecco medium (IMDM) supplemented with 15% FCS, 5% protein-free hybridoma medium (PFHM-II; Gibco), 2 mM l-glutamine (Cellgro), 0.5 mM ascorbic acid (Cellgro), 200 μg/mL transferrin (Roche), and 1.5 × 10−4 M MTG. For EBs harvested on day 3 or 4.5 of differentiation, ESCs were seeded at a density of 4 × 104 cells/mL culture medium, and for EBs harvested on day 5 or later, ESCs were seeded at a density of 8 × 103 cells/mL culture medium. For fluorescence-activated cell sorting, day 3 to 3.25 EBs were dissociated using Accumax and stained with anti-mouse Flk1–phycoerythrin (PE; both form eBioscience), collected into EB medium and recultured with equivalent numbers of dissociated day 2 EB-derived cells in low-cluster 6-well plates (Costar). For serum-free cultures, protocols were adapted from published reports.11,34 Briefly, ESCs were cultured in feeder-free 2i + LIF medium, then transferred to differentiation medium containing 75% IMDM + 25% Ham F12 supplemented with N2 (1:100) and B27 (1:50). On day 2, EBs were dissociated and recultured with vascular endothelial growth factor (VEGF) (5 ng/mL), BMP4 (2 ng/mL), and ActivinA (1 ng/mL). For analysis of effects downstream of BMP signaling, EBs were stimulated on day 4 with 12.5 ng/mL BMP4. Inhibitors were used as noted in the figure legends: LDN193189 or dorsomorphin (BMP inhibitors; Axon Medchem or Tocris), DAPT (N-[N-(3,5-difluorophenacetyl)-l-alanyl]-(S)-phenylglycine t-butyl ester, γ-secretase/NOTCH signaling inhibitor; Sigma), SCIO 469 (p38-α inhibitor; Tocris).
Inducible Smad5 (miSMAD5) single and Smad1/Smad5 double-knockdown cell lines (miSMAD51) were generated as described.33,35 Smad1-specific and Smad5-specific shRNA hairpins are noted in supplemental Table 1 (available on the Blood Web site). Multiple clonal lines were created and validated using methods previously described.33 Knockdown was achieved through induced expression of the hairpin RNA by addition to culture medium of 2 μg/mL doxycycline.
Hematopoietic colony assays
Differentiation of EB-derived cells into hematopoietic progenitor colonies was performed as described.33,36 Briefly, EBs were disaggregated on day 6 of differentiation, and single-cell suspensions were replated at a density of 1 × 105 cells/mL in IMDM containing 1% methylcellulose, 10% plasma-derived serum (PDS; Animal Technologies Inc), 5% PFHM-II, 2 mM l-glutamine, 0.25 mM ascorbic acid, and 1.5 × 10−4M MTG, supplemented with the following cytokines: primitive erythroblasts (human recombinant erythropoietin [EPO], 2 U/mL); macrophages (interleukin-3 [IL-3; 5 ng/mL], macrophage colony-stimulating factor [M-CSF; 5 ng/mL]); megakaryocytes (IL-3 [5 ng/mL], IL-11 [5 ng/mL], thrombopoietin [TPO], 5 ng/mL]); and mixed colonies (stem cell factor [SCF; 5 ng/mL], IL-3, granulocyte colony-stimulating factor [G-CSF; 30 ng/mL], granulocyte macrophage colony-stimulating factor [GM-CSF; 10 ng/mL], IL-11, IL-6 [5 ng/mL], TPO, M-CSF).
Colonies were counted after 4 days (erythroid progenitor [EryP]), 6 days (macrophage progenitor [MacP]), or 8 days (megakaryocyte progenitor [MegaP], Mixed). In control samples, colony numbers ranged between 800 and 2500 EryP, 80 and 250 MacP, and 40 and 200 MegaP per 105 cells. EPO was purchased from the Mount Sinai Medical Center hospital pharmacy. All other cytokines were purchased from R&D Systems.
Quantitative RT-PCR
Cells were harvested into TRIzol reagent on day 3.5 for total RNA extraction. For analysis of NOTCH ligands and C/EBPα, depletion or inhibition was performed on day 4 and cells were harvested on day 6 and day 8, respectively. Total RNA (1 μg) was subjected to reverse transcription (RT) reactions using Superscript III First-Strand reagent (Invitrogen) to generate complementary DNA (cDNA), which was diluted 1:25 in RNAse-free H2O for quantitative polymerase chain reaction (qPCR) analysis on a Roche 480 II Lightcycler using Sybr green detection and the 2−ΔΔCT method.37 Gene-specific qPCR primers are listed in supplemental Table 1.
Western blotting
Whole-cell extracts were collected from ESC or EB cultures in complete lysis buffer (20 mM Tris pH 7.5, 150 mM NaCl, 50 mM NaF, 1% NP-40 substitute, Halt protease inhibitor cocktail [Thermo Fisher, 1:100 dilution]). Proteins were resolved by electrophoresis on 10% NuPage Bis-Tris gels (Invitrogen) and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad). Membranes were blocked in 5% bovine serum albumin (BSA) in Tris-buffered saline (TBS) + 0.05% Tween 20 and probed overnight with the following antibodies: rabbit anti-SMAD1 XP, anti-phospho-SMAD1/5/9, anti-phospho-p38, anti-p38, anti-phospho-AKT, anti-pan AKT, anti-phospho-ERK1/2, anti-ERK1/2, anti-phospho-JNK, anti-JNK, anti-NOTCH intracellular domain (NICD), anti-phospho-SMAD2/3, anti-SMAD2/3 (all Cell Signaling), mouse anti-SMAD5 (Invitrogen), or mouse anti-β-Actin (Sigma). Proteins were visualized with horseradish peroxidase (HRP)–conjugated secondary antibodies (Bio-Rad) and West Pico chemiluminescence (Thermo Fisher).
Results
Conditional, simultaneous depletion of SMAD5 returns hematopoietic potential to homeostatic levels in SMAD1-depleted EBs
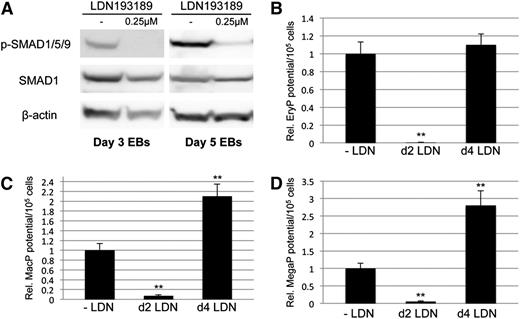
We showed previously, using conditional Smad1 depletion, that Smad1 normally restricts both erythroid and myeloid potential around day 4 of EB development.33 Here we tested the activity of the related Smad5 in this context. For this purpose, ESC lines were generated, similar to the previous Smad1 knockdown line (miSMAD1), using the AinV system for conditional depletion of Smad5 alone or together with Smad1. These ESC lines are designated miSMAD5 and miSMAD51, respectively. Targeting of Smad1 and Smad5 together was achieved by recombining tandem gene-specific shRNA hairpins into a single tetracycline-responsive, engineered site on the mouse X chromosome in the AinV18 parental line of ESCs.33 AinV18 ESCs express the reverse tet transactivator under control of the Rosa26 locus, so that transgene (or in this case, hairpin) expression can be induced via doxycycline (dox) treatment.38 The targeted transgene includes a cassette for internal ribosomal entry site (IRES)–driven expression of enhanced green fluorescent protein (GFP), and expression is abrogated by dox withdrawal, allowing temporal control of gene expression in differentiating EBs. Cell extracts from dox-treated EBs, including those derived from miSMAD1 ESCs,33 were subjected to western blotting analysis of SMAD5 and SMAD1 protein levels (Figure 1A), which demonstrated that the relevant targeted proteins were knocked down at steady-state at least 50% in EBs treated with 2 μg/mL dox (Figure 1B). Consequently, SMAD1/5/9 activation was reduced to a similar extent (supplemental Figure 1). This is consistent with the published Smad1-specific conditional knockdown,33 which was previously shown to block hematopoietic mesoderm when induced at day 2, but to enhance hematopoietic progenitor output when induced at day 4.
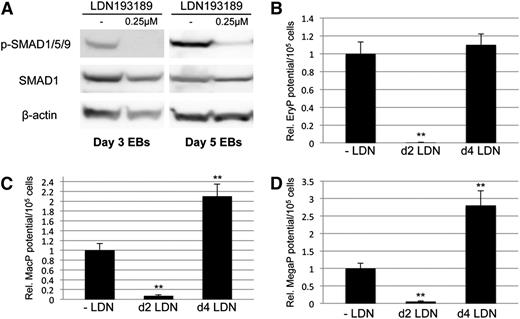
Conditional depletion of Smad1 and Smad5 reveals differing contributions of each to primitive hematopoiesis. Cre/loxP-mediated homologous recombination was used to target insertion of constructs containing shRNA hairpins corresponding to sequences within the MH1 domain of Smad1 alone (miSMAD1), Smad5 alone (miSMAD5), or both Smad5 and Smad1 (miSMAD51). Each hairpin construct is targeted to the identical site downstream of an operator/promoter, which allows tet-on activation via dox treatment (miSMAD1 was previously characterized33 ). (A) Representative western blotting analysis of SMAD5 and SMAD1 protein expression in lysates derived from EBs after 24 hours of dox treatment (DOX, compared with uninduced control). (B) Quantitative densitometric analysis of samples induced with 2 μg/mL dox normalized to control β-actin levels from at least 3 separate experiments to derive mean ± SEM. (C-E) Embryoid bodies derived from parental AinV18 cells, miSMAD5, miSMAD51, or miSMAD1 ESCs were induced with dox on day 2 (dox d2) or day 4 (dox d4) of EB differentiation and evaluated by hematopoietic colony assays to measure primitive erythroid (EryP [C]) and myeloid (macrophages, MacP [D]; megakaryocytes, MegaP [E]) potential. Colony assays were each performed at least 3 separate times, with samples counted in triplicate, and values are reported as normalized to untreated controls. Statistical significance compared with no dox controls, with *P < .05 and **P < .01, respectively. CTL, control; dox, doxycycline.
Conditional depletion of Smad1 and Smad5 reveals differing contributions of each to primitive hematopoiesis. Cre/loxP-mediated homologous recombination was used to target insertion of constructs containing shRNA hairpins corresponding to sequences within the MH1 domain of Smad1 alone (miSMAD1), Smad5 alone (miSMAD5), or both Smad5 and Smad1 (miSMAD51). Each hairpin construct is targeted to the identical site downstream of an operator/promoter, which allows tet-on activation via dox treatment (miSMAD1 was previously characterized33 ). (A) Representative western blotting analysis of SMAD5 and SMAD1 protein expression in lysates derived from EBs after 24 hours of dox treatment (DOX, compared with uninduced control). (B) Quantitative densitometric analysis of samples induced with 2 μg/mL dox normalized to control β-actin levels from at least 3 separate experiments to derive mean ± SEM. (C-E) Embryoid bodies derived from parental AinV18 cells, miSMAD5, miSMAD51, or miSMAD1 ESCs were induced with dox on day 2 (dox d2) or day 4 (dox d4) of EB differentiation and evaluated by hematopoietic colony assays to measure primitive erythroid (EryP [C]) and myeloid (macrophages, MacP [D]; megakaryocytes, MegaP [E]) potential. Colony assays were each performed at least 3 separate times, with samples counted in triplicate, and values are reported as normalized to untreated controls. Statistical significance compared with no dox controls, with *P < .05 and **P < .01, respectively. CTL, control; dox, doxycycline.
When Smad5 expression is depleted in EBs at day 2 there is no phenotype, indicating that only Smad1 is required for early mesoderm patterning. However, when knockdown is induced at day 4, there is a specific decrease in primitive erythroid potential (Figure 1C), opposite of Smad1 knockdown phenotype, and there is no change in primitive myeloid potential (Figure 1D-E). These observations are consistent with our previous results in zebrafish.39 When Smad1/Smad5 expression is simultaneously depleted during the early stages of germ layer commitment, there is a general failure to specify hematopoietic progenitors in comparison with untreated controls (Figure 1C-E), equivalent to knockdown of Smad1 alone. However, while Smad1 depletion starting at day 4 of EB differentiation enhances hematopoietic output,33 codepletion of Smad5 and Smad1 at day 4 dropped hematopoietic capacity back to approximately control levels (Figure 1C-E). This suggests that Smad5 activity is responsible for the expanded hematopoietic fate under conditions of limited Smad1.
Inhibition of BMP signaling in EB cultures specifically promotes primitive myeloid potential
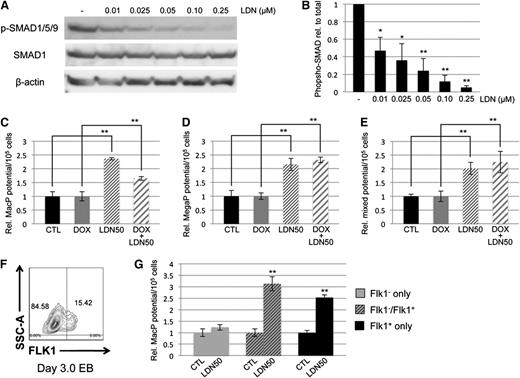
To confirm our interpretation that hematopoietic potential is balanced by levels of Smad1/Smad5, we used an alternative approach to block function by targeting BMP receptor signaling upstream of R-SMAD activation, and examining whether BMP inhibition is phenotypically equivalent to Smad1/Smad5 knockdown. For this purpose, we used the small-molecule inhibitor LDN193189 (LDN), a compound that specifically blocks activation of ALK2/ALK3 BMP type I receptors.40 EB cultures were treated for 24 hours with LDN starting on day 2 or day 4 of differentiation. Concentrations of LDN as low as 250 nM were sufficient to essentially eliminate BMP signaling, as measured by serine phosphorylation of SMAD1/5/9 (Figure 2A). As with Smad1 depletion, or Smad1/5 depletion, early BMP inhibition at day 2 caused failure of hematopoietic potential in methylcellulose-based progenitor colony assays measuring primitive erythroid (EryP, Figure 2B), macrophage (MacP, Figure 2C), and megakaryocyte (MegaP, Figure 2D) progenitor populations, each of which decreased at least 20-fold with respect to untreated controls. Similar to co-depletion of Smad1/Smad5, later inhibition of BMP signaling with LDN on day 4 of EB differentiation did not affect erythroid potential (Figure 2B). These observations were expected outcomes of blocking pathway-restricted R-SMAD signaling, whether by directly targeting Smad1/Smad5 or inhibiting upstream BMP receptor activation. Unexpectedly, however, following day 4 LDN treatment we noted a significant increase of approximately twofold to threefold in hematopoietic progenitor potential specific to the primitive myeloid lineages (Figure 2C-D).
Small-molecule–based inhibition of BMP signaling results in increased primitive myeloid potential. (A) Murine EBs were treated for 24 hours with 0.25 μM LDN, harvested on the indicated days of EB differentiation, and subjected to western blotting analysis of pathway-restricted R-SMAD phosphorylation. Phospho-SMAD levels were compared with total expression of SMAD1, and β-actin was used as a loading control. Embryoid bodies treated with LDN on day 2 or 4 of EB differentiation were cultured to day 6, harvested, and seeded on cytokine-supplemented methylcellulose medium to gauge primitive hematopoietic potential, including (B) erythroid progenitors and early myeloid lineages, (C) macrophage progenitors, and (D) megakaryocyte progenitors. Results of hematopoietic progenitor assays are reported as mean ± SEM from at least 3 experimental repeats, with values normalized to relevant controls. **P < .01. EryP, erythroid progenitor; MacP, macrophage progenitor; MegaP, megakaryocyte progenitor.
Small-molecule–based inhibition of BMP signaling results in increased primitive myeloid potential. (A) Murine EBs were treated for 24 hours with 0.25 μM LDN, harvested on the indicated days of EB differentiation, and subjected to western blotting analysis of pathway-restricted R-SMAD phosphorylation. Phospho-SMAD levels were compared with total expression of SMAD1, and β-actin was used as a loading control. Embryoid bodies treated with LDN on day 2 or 4 of EB differentiation were cultured to day 6, harvested, and seeded on cytokine-supplemented methylcellulose medium to gauge primitive hematopoietic potential, including (B) erythroid progenitors and early myeloid lineages, (C) macrophage progenitors, and (D) megakaryocyte progenitors. Results of hematopoietic progenitor assays are reported as mean ± SEM from at least 3 experimental repeats, with values normalized to relevant controls. **P < .01. EryP, erythroid progenitor; MacP, macrophage progenitor; MegaP, megakaryocyte progenitor.
We showed previously that conditional depletion of Smad1 expression early during the germ layer commitment stages of EB differentiation impairs mesoderm formation.33 As expected, either BMP inhibition with LDN or Smad1/Smad5 co-knockdown at this stage caused downregulation of mesoderm and primitive streak marker gene transcripts, including Brachyury, Evx1, HoxB1, Mesp1, Mesp2, Goosecoid, and MixL1 (supplemental Figure 2A-B). However, the consequence of Smad1/5 depletion is clearly different from blocking BMP signaling with LDN, as it resulted in upregulation of endodermal transcripts, while in contrast LDN pushed cultures toward neural ectoderm fates (supplemental Figure 2C-D), consistent with previous reports.33,41,42 This observation most likely reflects the fact that LDN, in addition to blocking R-SMAD activation, will modulate SMAD-independent downstream effects.
SMAD-independent BMP-mediated regulation of myeloid potential
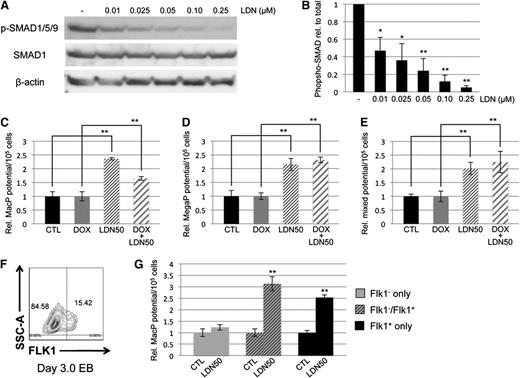
Because the enhanced myeloid phenotype was not observed in the Smad1/Smad5 co-knockdown, and because expression of Smad9 transcripts was comparatively very low (data not shown), we hypothesized that BMP control of primitive myeloid potential was SMAD-independent. One caveat is that the Smad1/Smad5 shRNA-mediated co-knockdown was not as complete compared with the loss of p-SMAD1/5/9 achieved with LDN. Therefore, LDN was titrated in day 4 EB cultures to determine the concentration necessary to inhibit BMP signaling ∼50%, resulting in conditions analogous to the partial depletion of Smad1/Smad5 expression in the dox-induced miSMAD51-derived EBs. Western blotting analysis of SMAD1/5/9 phosphorylation displayed a linear dose-response relationship from complete inhibition at 250 nM to 50% inhibition at 10 nM (Figure 3A); this treatment is hereafter referred to as LDN50 in the figures and text to distinguish it from complete inhibition, referred to as LDN. Using LDN50, the potential of cultures to generate primitive macrophage, megakaryocyte, and mixed lineage progenitors was examined and found to recapitulate the previous results with LDN treatment (Figure 3C-E). Specifically, 50% inhibition of BMP signaling with LDN50 was sufficient to promote a significant increase in myeloid potential of approximately twofold to threefold over untreated controls. In samples cotreated with dox and LDN50 to simultaneously deplete Smad1/Smad5 and inhibit upstream signaling, there was again a significant increase in myeloid potential compared with control samples treated with dox only. This effect cannot be rescued by SMAD1, as LDN50 treatment of a conditional SMAD1-expressing ESC line32 resulted in increased myeloid potential, either with or without induction of SMAD1 (supplemental Figure 3A-B). Early hematovascular progenitors were collected from differentiating EBs by fluorescence-activated cell sorting the initial Flk1+ cell population on days 3 to 3.25 of differentiation (Figure 3F), and reculturing them in the presence of LDN50 to examine myeloid potential (Figure 3G). A similar relative increase in primitive macrophage colonies was observed compared with whole EB cultures, confirming that the effect of blocking BMP for myeloid progenitor potential is specific for this progenitor subset.
Control of myeloid potential by BMP signaling is SMAD-independent. (A) Titration of LDN compound and western blotting analysis of SMAD1/5/9 phosphorylation compared with total SMAD1 expression shows dose dependence of BMP signaling in day 4/5 EBs, with ∼50% inhibition achieved under conditions of 0.01 μM LDN (B, henceforth noted as LDN50). Analysis of myeloid potential by methylcellulose colony assays to examine (C) macrophage, (D) megakaryocyte, and (E) mixed lineage progenitors shows persistence of myeloid phenotype after LDN treatment regardless of whether baseline is untreated control (black bar compared with thin-striped bar) or dox-induced Smad5/Smad1 knockdown (gray bar compared with wide-striped bar). (F) Effects of BMP signaling inhibition on myeloid potential were examined on progenitors by sorting Flk1+ cells from day 3 EBs, and reculturing them 24 hours later in the presence of LDN50. Shown is a representative sort. (G) Macrophage potential was measured; gray bars correspond to FLK1− cells, striped bars to a 50/50 mix of FLK1− and FLK1+, and black bars to cultures containing FLK1+ only. Colony assays were each performed at least 3 separate times, with samples counted in triplicate. *P < .05; **P < .01.
Control of myeloid potential by BMP signaling is SMAD-independent. (A) Titration of LDN compound and western blotting analysis of SMAD1/5/9 phosphorylation compared with total SMAD1 expression shows dose dependence of BMP signaling in day 4/5 EBs, with ∼50% inhibition achieved under conditions of 0.01 μM LDN (B, henceforth noted as LDN50). Analysis of myeloid potential by methylcellulose colony assays to examine (C) macrophage, (D) megakaryocyte, and (E) mixed lineage progenitors shows persistence of myeloid phenotype after LDN treatment regardless of whether baseline is untreated control (black bar compared with thin-striped bar) or dox-induced Smad5/Smad1 knockdown (gray bar compared with wide-striped bar). (F) Effects of BMP signaling inhibition on myeloid potential were examined on progenitors by sorting Flk1+ cells from day 3 EBs, and reculturing them 24 hours later in the presence of LDN50. Shown is a representative sort. (G) Macrophage potential was measured; gray bars correspond to FLK1− cells, striped bars to a 50/50 mix of FLK1− and FLK1+, and black bars to cultures containing FLK1+ only. Colony assays were each performed at least 3 separate times, with samples counted in triplicate. *P < .05; **P < .01.
As a control for specificity, we tested an alternative BMP signaling inhibitor, dorsomorphin. BMP signaling in day 4 EBs was ∼10-fold less sensitive to dorsomorphin treatment (supplemental Figure 4A-B), but similar effects in myeloid potential were observed after 50% inhibition, showing significant twofold to threefold increases in macrophage and megakaryocyte progenitors based on colony assays (supplemental Figure 4C-D).
SMAD-independent BMP control of myeloid potential requires an intact NOTCH signaling pathway
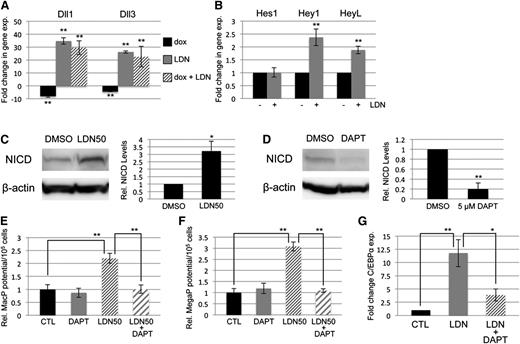
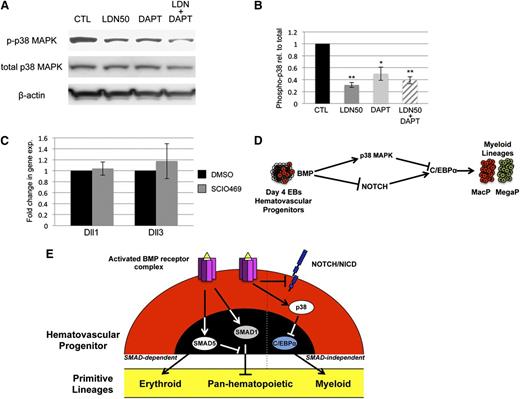
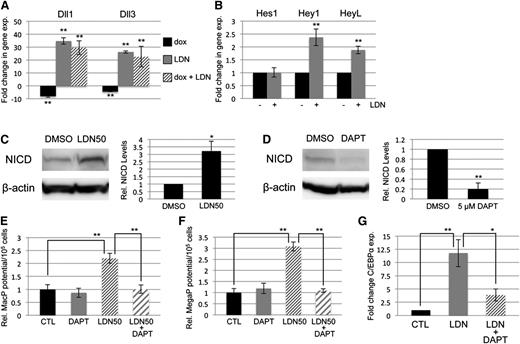
BMP signaling has been reported by several groups to cooperate with both NOTCH and WNT signaling in its control of early mesoderm commitment and hematovascular development,24-26 so we profiled transcript levels encoding a wide range of ligands and receptors in both pathways by quantitative RT-PCR analysis (qPCR, not shown). Transcripts encoding 2 ligands of the NOTCH pathway, Delta-like protein 1 and 3 (Dll1 and Dll3), were significantly upregulated after BMP inhibition with LDN, either with or without depletion of Smad1/Smad5 (Figure 4A). In contrast, Smad1/Smad5 depletion alone resulted in a smaller but opposite effect, causing downregulation of Dll1 and Dll3 expression (Figure 4A). To assess whether BMP inhibition has bona fide effects on NOTCH signaling, we examined expression levels of NOTCH effector genes (Figure 4B), and found significant increases in transcript levels for Hey1 and HeyL. Inhibition with LDN50 resulted in an approximately threefold increase in NOTCH receptor activation, as measured by western blotting analysis of intracellular domain accumulation (NICD, Figure 4C).
SMAD-independent BMP control of myeloid potential is dependent on NOTCH, associated with increased expression of Δ-like ligands. (A) Quantitative RT-PCR analysis of Δ-like genes Dll1 and Dll3 in day 5 EBs after 24-hour treatment with dox to deplete Smad5 and Smad1 (black bar); LDN only to inhibit BMP signaling (gray bar); or dox + LDN demonstrating that enhanced expression is SMAD-independent (striped bar). (B) qPCR analysis of NOTCH effector genes 24 hours after BMP inhibition with LDN. (C) Western blotting analysis of changes in NOTCH activation via cleaved receptor intracellular domain levels (NICD), 24 hours after BMP inhibition by LDN50 in day 4 EBs. (D) Western blotting analysis of NICD 24 hours after 5 μM DAPT treatment of day 4 EBs. Representative blots are shown with densitometric analysis of relative levels compared with untreated controls. Methylcellulose colony assay analyses of (E) macrophage and (F) megakaryocyte potential under conditions of LDN50 treatment alone (compare small striped gray bars to black control bars) or in combination with 5 μM DAPT (compare gray bars to large striped bars), with each experiment performed at least 3 separate times. (G) qPCR analysis of transcript levels for myeloid regulatory transcription factor C/EBPα on day 8 of EB differentiation after day 4 treatment with LDN alone (compare black control bar to gray bar), or cotreatment with LDN + 5 μM DAPT (compare gray bar to striped bar). Values in qPCR assays are reported as mean ± SEM from at least 3 experimental repeats, normalized to untreated controls, and compared with Gapdh as a reference gene. In all experiments, *P < .05 and **P < .01.
SMAD-independent BMP control of myeloid potential is dependent on NOTCH, associated with increased expression of Δ-like ligands. (A) Quantitative RT-PCR analysis of Δ-like genes Dll1 and Dll3 in day 5 EBs after 24-hour treatment with dox to deplete Smad5 and Smad1 (black bar); LDN only to inhibit BMP signaling (gray bar); or dox + LDN demonstrating that enhanced expression is SMAD-independent (striped bar). (B) qPCR analysis of NOTCH effector genes 24 hours after BMP inhibition with LDN. (C) Western blotting analysis of changes in NOTCH activation via cleaved receptor intracellular domain levels (NICD), 24 hours after BMP inhibition by LDN50 in day 4 EBs. (D) Western blotting analysis of NICD 24 hours after 5 μM DAPT treatment of day 4 EBs. Representative blots are shown with densitometric analysis of relative levels compared with untreated controls. Methylcellulose colony assay analyses of (E) macrophage and (F) megakaryocyte potential under conditions of LDN50 treatment alone (compare small striped gray bars to black control bars) or in combination with 5 μM DAPT (compare gray bars to large striped bars), with each experiment performed at least 3 separate times. (G) qPCR analysis of transcript levels for myeloid regulatory transcription factor C/EBPα on day 8 of EB differentiation after day 4 treatment with LDN alone (compare black control bar to gray bar), or cotreatment with LDN + 5 μM DAPT (compare gray bar to striped bar). Values in qPCR assays are reported as mean ± SEM from at least 3 experimental repeats, normalized to untreated controls, and compared with Gapdh as a reference gene. In all experiments, *P < .05 and **P < .01.
To determine whether NOTCH pathway activation functionally impacts progenitor output, NOTCH signaling was blocked by treatment with the γ-secretase inhibitor DAPT (Figure 4D). As shown in Figure 4E-F, day 4 treatment with 5 μM DAPT alone had no effect on progenitor output, but when added with LDN50 it abrogated the increase in myeloid potential observed with LDN50 treatment alone, reducing MacP and MegaP colony numbers to control levels. Additionally, qPCR analysis showed that LDN-induced inhibition of BMP signaling resulted in specific upregulation of the myeloid-regulatory transcription factor C/EBPα, which was also largely dependent on functional NOTCH signaling (Figure 4G).
SMAD-independent BMP control of myeloid potential proceeds through the p38MAPK pathway
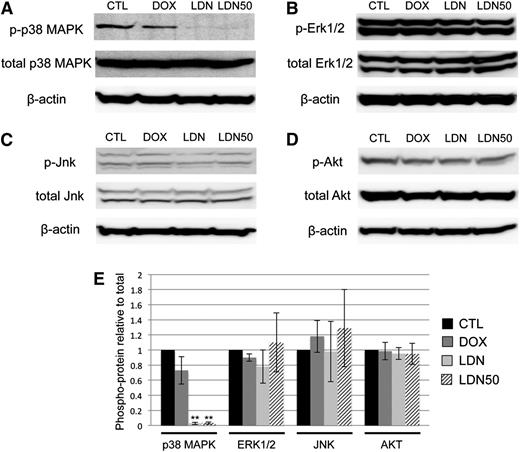
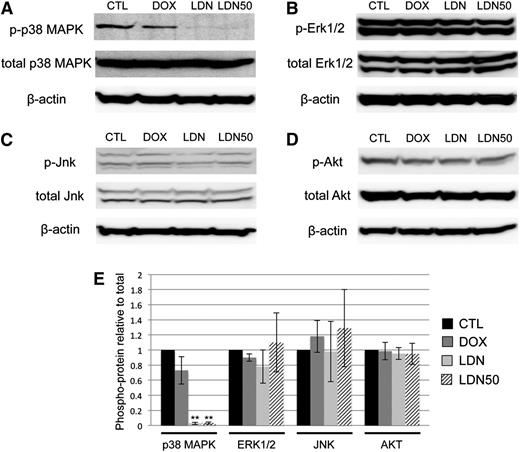
Because BMP control of myeloid potential in EBs occurred independent of SMAD signaling, we sought to determine the specific downstream pathway(s) responsible. We reasoned that activation of the putative pathway would be inhibited to some extent by LDN and/or LDN50 treatment, but not by dox-induced Smad1/Smad5 knockdown. Indeed, we found that LDN or LDN50 treatment of day 4 EBs specifically resulted in marked deactivation of p38MAPK signaling, as shown by western blotting analysis of phospho(Thr/Tyr)-p38 (Figure 5A). Any changes in baseline p38MAPK activity in dox-treated controls were not statistically significant. In contrast, other candidate signaling pathways were unchanged regardless of treatment, including the ERK1/2 (Figure 5B) and JNK (Figure 5C) branches of MAPK signaling, as well as the PI3K-AKT signaling pathway (Figure 5D-E). Moreover, LDN-mediated inhibition of BMP signaling did not result in a significant shift to TGF-β pathway-restricted SMAD activation, measured by SMAD2/3 phosphorylation levels (supplemental Figure 5).
BMP inhibition by LDN is associated with deactivation of p38MAPK. Phosphorylation states of reported downstream kinases responsive to SMAD-independent BMP signaling were probed in western blotting experiments and compared with total protein levels with β-actin as a loading control. (A) p38MAPK Thr/Tyr phosphorylation was markedly downregulated after overnight treatment with either full-dose LDN or LDN50 to achieve ∼100% or 50% BMP signaling inhibition, respectively, with co-depletion of SMAD1/5 by dox induction resulting in no significant changes in p38 phosphorylation. In contrast, (B) Thr/Tyr phosphorylation of ERK1/2, (C) Thr/Tyr phosphorylation of JNK, and (D) Ser phosphorylation of AKT displayed no significant changes compared with controls for any treatment. (E) These relationships are presented as quantified densitometry data, where **P < .01.
BMP inhibition by LDN is associated with deactivation of p38MAPK. Phosphorylation states of reported downstream kinases responsive to SMAD-independent BMP signaling were probed in western blotting experiments and compared with total protein levels with β-actin as a loading control. (A) p38MAPK Thr/Tyr phosphorylation was markedly downregulated after overnight treatment with either full-dose LDN or LDN50 to achieve ∼100% or 50% BMP signaling inhibition, respectively, with co-depletion of SMAD1/5 by dox induction resulting in no significant changes in p38 phosphorylation. In contrast, (B) Thr/Tyr phosphorylation of ERK1/2, (C) Thr/Tyr phosphorylation of JNK, and (D) Ser phosphorylation of AKT displayed no significant changes compared with controls for any treatment. (E) These relationships are presented as quantified densitometry data, where **P < .01.
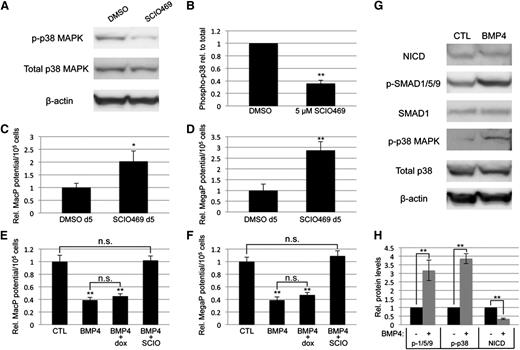
To confirm that myeloid progenitor capacity is modulated by BMP signaling through p38MAPK activity, we directly targeted p38MAPK with a small-molecule inhibitor highly specific to the p38-α isoform, SCIO469, which significantly attenuated phosphorylation in EB cultures (Figure 6A-B). Treatment on day 5 of EB differentiation, at a time point corresponding to events downstream of BMP activation, resulted in enhanced myeloid progenitor output, similar to LDN50-based BMP inhibition, with significant (twofold to threefold) increases in macrophage (Figure 6C) and megakaryocyte potential (Figure 6D). Finally, we examined the outcome of adding recombinant BMP4 under defined conditions in serum-free cultures. Stimulation on day 4 with BMP4 resulted in twofold to threefold attenuation of myeloid potential (Figure 6E-F), inversely consistent with experiments using LDN (Figure 2C-D). As expected, BMP treatment activated SMAD1/5/9 signaling; additionally, it resulted in significant activation of p38MAPK and reduction of Notch signaling (Figure 6G-H), again the inverse result of LDN treatment in serum-containing cultures (Figures 4C and 5A).
Direct inhibition of p38MAPK signaling is sufficient to control primitive myeloid potential. (A) Western blotting analysis of total and phospho-p38 levels after treatment of day 5 EBs with DMSO (vehicle control) or SCIO469, a small-molecule inhibitor specific to the p38-α isoform. (B) Densitometric analysis from 3 separate experiments, with phospho-p38 normalized to total p38 protein expression levels. Primitive myeloid potential was analyzed using methylcellulose-based progenitor colony assays, which showed that (C) MacP and (D) MegaP potential after direct p38MAPK inhibition is enhanced to levels similar to those observed after BMP inhibition. Macrophage (E) and megakaryocyte (F) potential was analyzed in serum-free EB cultures after addition at day 4 of recombinant BMP4, with or without the p38-α inhibitor SCIO469. BMP4 treatment directly attenuated myeloid potential and this was rescued by simultaneous p38 inhibition. Furthermore, BMP4 increased p38 activation and decreased NOTCH pathway signaling, measured by western blotting analysis of phospho-p38 and cleaved NOTCH intracellular domain, respectively (G). Western blotting results are quantified from 3 separate experiments in panel H. Values are normalized to DMSO-treated (A-F) or untreated (G-H) controls; asterisks indicate significance, with *P < .05 and **P < .01, respectively. DMSO, dimethylsulfoxide.
Direct inhibition of p38MAPK signaling is sufficient to control primitive myeloid potential. (A) Western blotting analysis of total and phospho-p38 levels after treatment of day 5 EBs with DMSO (vehicle control) or SCIO469, a small-molecule inhibitor specific to the p38-α isoform. (B) Densitometric analysis from 3 separate experiments, with phospho-p38 normalized to total p38 protein expression levels. Primitive myeloid potential was analyzed using methylcellulose-based progenitor colony assays, which showed that (C) MacP and (D) MegaP potential after direct p38MAPK inhibition is enhanced to levels similar to those observed after BMP inhibition. Macrophage (E) and megakaryocyte (F) potential was analyzed in serum-free EB cultures after addition at day 4 of recombinant BMP4, with or without the p38-α inhibitor SCIO469. BMP4 treatment directly attenuated myeloid potential and this was rescued by simultaneous p38 inhibition. Furthermore, BMP4 increased p38 activation and decreased NOTCH pathway signaling, measured by western blotting analysis of phospho-p38 and cleaved NOTCH intracellular domain, respectively (G). Western blotting results are quantified from 3 separate experiments in panel H. Values are normalized to DMSO-treated (A-F) or untreated (G-H) controls; asterisks indicate significance, with *P < .05 and **P < .01, respectively. DMSO, dimethylsulfoxide.
BMP controls primitive myeloid potential in parallel through NOTCH and p38MAPK pathways
To probe epistatic relationships between p38 and NOTCH, we examined NOTCH inhibition in the context of p38 signaling and vice versa. As shown in Figure 7A-B, DAPT treatment of EB cultures results in some deactivation of p38MAPK, although not to the same extent as BMP inhibition with LDN50. Moreover, DAPT alone did not alter myeloid potential (Figure 4E-F). Under conditions of p38 inhibition with SCIO469, expression of Dll1 and Dll3 transcripts did not change (Figure 7C). Therefore, there does not appear to be functional cross-talk between the NOTCH and p38 pathways. These results are consistent with parallel functions for NOTCH and p38 downstream of BMP, as schematized in Figure 7D.
NOTCH and p38 function in parallel pathways to balance primitive myelopoiesis. (A) p38MAPK activation was examined in day 5 EBs by western blotting experiments under conditions of LDN50 treatment alone or in combination with 5 μM DAPT treatment, with β-actin as loading control. (B) Phospho-p38 levels were analyzed in comparison with total p38, and reported as mean ± SEM from 3 separate experiments. *P < .05 and **P < .01, respectively. Although DAPT alone decreases pp38 levels, it is not as significant as LDN, and does not impact myeloid output in the absence of LDN. (C) qPCR analysis of Dll1 and Dll3 Notch ligand genes after p38 inhibition with SCIO469. (D) Model diagram of BMP signaling control of myeloid potential proposing parallel pathways that function downstream through NOTCH and p38MAPK and converge to balance C/EBP levels. (E) The schematic summarizes ways that BMP signaling modulates primitive hematopoiesis through both SMAD-dependent and SMAD-independent mechanisms. Based on ESC differentiation studies, SMAD1 normally functions in hematovascular precursors to generally restrict myeloid and erythroid (panhematopoietic) potential. SMAD5 normally balances this effect while at the same time uniquely promoting primitive erythroid potential. BMP receptor activation separately functions to restrict C/EBPα-regulated myeloid capacity, independent of SMAD activity, through p38MAPK activation and Notch pathway inhibition.
NOTCH and p38 function in parallel pathways to balance primitive myelopoiesis. (A) p38MAPK activation was examined in day 5 EBs by western blotting experiments under conditions of LDN50 treatment alone or in combination with 5 μM DAPT treatment, with β-actin as loading control. (B) Phospho-p38 levels were analyzed in comparison with total p38, and reported as mean ± SEM from 3 separate experiments. *P < .05 and **P < .01, respectively. Although DAPT alone decreases pp38 levels, it is not as significant as LDN, and does not impact myeloid output in the absence of LDN. (C) qPCR analysis of Dll1 and Dll3 Notch ligand genes after p38 inhibition with SCIO469. (D) Model diagram of BMP signaling control of myeloid potential proposing parallel pathways that function downstream through NOTCH and p38MAPK and converge to balance C/EBP levels. (E) The schematic summarizes ways that BMP signaling modulates primitive hematopoiesis through both SMAD-dependent and SMAD-independent mechanisms. Based on ESC differentiation studies, SMAD1 normally functions in hematovascular precursors to generally restrict myeloid and erythroid (panhematopoietic) potential. SMAD5 normally balances this effect while at the same time uniquely promoting primitive erythroid potential. BMP receptor activation separately functions to restrict C/EBPα-regulated myeloid capacity, independent of SMAD activity, through p38MAPK activation and Notch pathway inhibition.
Discussion
By blocking type I BMP receptors with the antagonist LDN193189, we discovered that in post-hemangioblast stage EB cultures, BMP signaling restricts generation of myeloid precursors. Myeloid progenitor numbers increase under conditions of BMP inhibition with LDN (or dorsomorphin) whether or not R-SMAD effectors are modulated. We found that inhibition of BMP with LDN (but not by SMAD knockdown) causes a marked upregulation of NOTCH signaling molecules, specifically Delta-like ligands, and activates NOTCH, assessed by generation of the processed transcription factor NICD. Inhibition of NOTCH signaling in the presence of LDN restores C/EBPα expression levels and myeloid progenitor numbers back to control levels. Finally, we found that at this stage of EB development SMAD-independent BMP signaling proceeds preferentially through activation of the p38 branch of MAPK signaling.
There are caveats to our interpretation of a SMAD-independent role for BMP inhibition by LDN. First, knockdown of Smad1/Smad5 using inducible hairpins is not complete. However, knockdown was adequate to achieve reproducible hematopoietic phenotypes in both Smad1/Smad5 knockdown and BMP inhibition models. In addition, these levels of depletion are sufficient to significantly disrupt earlier mesoderm-generating steps around day 2 of EB development. Using LDN50 conditions we confirmed that 50% inhibition of BMP signaling is sufficient to affect downstream gene expression and myeloid progenitor colony formation. Second, our experiments have excluded Smad1 and Smad5 from mediating the LDN effect, but we cannot exclude a potential function for the related pathway-restricted R-SMAD, Smad9. However, we found it difficult to reliably detect significant Smad9 transcript levels in this system, suggesting that Smad9 is unlikely to contribute to the observed phenotypes.
We also discovered Smad1/5-independent roles for BMP signaling in EB cultures prior to hematopoietic commitment. Although we did not explore the mechanism, day 2 treatment of EBs with LDN pushes cultures toward neural ectoderm fates including upregulation of Pax6. This differs in phenotype from day 2 codepletion of Smad1/Smad5, which results in a bias toward endoderm (supplemental Figure 1). Consistent with this observation, a similar neural ectoderm fate was reported for hESC-derived differentiating cultures inhibited for BMP signaling using either NOGGIN41 or chemical antagonists.42
During stages of hematopoietic commitment, we showed that blocking BMP signaling altered 2 SMAD-independent pathways, causing enhanced NOTCH signaling, and decreased p38 activation. That these changes are dependent on BMP was confirmed using serum-free defined culture conditions. Adding recombinant BMP4 resulted in the inverse phenotype compared with LDN treatment, namely a reduction in myeloid potential with concomitantly increased p38MAPK activity and reduced NOTCH pathway signaling. NOTCH could directly suppress p38 activation through induction of a p38 phosphatase, as has been shown in the context of myogenesis.43 Indeed, LDN treatment caused modest activation of the phosphatase DUSP1 (not shown). However, this may not be functionally relevant, as blocking NOTCH with DAPT led to a partial decrease of phospho-p38, rather than activating p38 (Figure 7), and has no effect on steady state myeloid progenitor output. Rather, the requirement for NOTCH to balance myeloid output is only revealed under conditions of LDN treatment, when phospho-p38 levels are significantly depleted, and NOTCH activity is enhanced. We also saw no effect on expression of NOTCH ligands when p38 activation is blocked. Therefore, the data are most consistent with parallel NOTCH and p38 pathways, activating and restricting C/EBPα transcription levels, respectively, in order to balance the myeloid progenitor output (Figure 7D). In T cells, NOTCH was shown to repress C/EBPα levels through activation of the repressor HES1.44 In our system probing primitive myelopoiesis, NOTCH activation is in contrast associated with enhanced C/EBPα levels. Hes1 levels are not affected, but the alternative NOTCH targets Hey1 and HeyL are enhanced, and may impact C/EBP indirectly by repressing factors that normally limit C/EBPα expression. Various aspects of this regulatory network may function non-cell-autonomously, which is an added level of complexity our study did not distinguish. The effects downstream of LDN are at least autonomous to the early subset of FLK1+ progenitors, although we cannot rule out cross-talk among cells within this progenitor set.
Regarding regulation of p38MAPK by BMP signaling, we considered the TGF-β–responsive TAK1 kinase.45 Activation of TAK1 was unaffected by treatment with LDN in day 4 EB cultures (data not shown). Incorporating results from this and 2 previous studies manipulating BMP signaling in the ESC/EB system reveals distinct SMAD-dependent and SMAD-independent effects of BMP signaling in the control of primitive hematopoietic potential, with Smad5 being required for normal erythroid potential, as well as counteracting Smad1 to balance homeostasis in all primitive lineages. These functions are separate, and can be examined in isolation, from SMAD-independent control of myeloid potential through NOTCH and p38MAPK signaling (summarized in Figure 7E). It may be feasible to exploit these findings from ESC differentiation culture systems to manipulate the generation and balance of hematopoietic stem and progenitor cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ting-Chun Liu for valuable technical contributions and Karen Mendelson for critiquing the manuscript.
This work was supported by National Institutes of Health, National Heart, Lung, and Blood Institute grant R37HL056182 (T.E.) and National Institute of Diabetes and Digestive and Kidney Diseases grant K01-KD096031 (B.D.C.).
Authorship
Contribution: B.D.C. conceived the study, carried out the experiments, and wrote the manuscript; and T.E. conceived the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Todd Evans, Department of Surgery, Weill Cornell Medical College, 1300 York Ave, LC-708, New York, NY 10065; e-mail: tre2003@med.cornell.edu.

![Figure 1. Conditional depletion of Smad1 and Smad5 reveals differing contributions of each to primitive hematopoiesis. Cre/loxP-mediated homologous recombination was used to target insertion of constructs containing shRNA hairpins corresponding to sequences within the MH1 domain of Smad1 alone (miSMAD1), Smad5 alone (miSMAD5), or both Smad5 and Smad1 (miSMAD51). Each hairpin construct is targeted to the identical site downstream of an operator/promoter, which allows tet-on activation via dox treatment (miSMAD1 was previously characterized33). (A) Representative western blotting analysis of SMAD5 and SMAD1 protein expression in lysates derived from EBs after 24 hours of dox treatment (DOX, compared with uninduced control). (B) Quantitative densitometric analysis of samples induced with 2 μg/mL dox normalized to control β-actin levels from at least 3 separate experiments to derive mean ± SEM. (C-E) Embryoid bodies derived from parental AinV18 cells, miSMAD5, miSMAD51, or miSMAD1 ESCs were induced with dox on day 2 (dox d2) or day 4 (dox d4) of EB differentiation and evaluated by hematopoietic colony assays to measure primitive erythroid (EryP [C]) and myeloid (macrophages, MacP [D]; megakaryocytes, MegaP [E]) potential. Colony assays were each performed at least 3 separate times, with samples counted in triplicate, and values are reported as normalized to untreated controls. Statistical significance compared with no dox controls, with *P < .05 and **P < .01, respectively. CTL, control; dox, doxycycline.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/3/10.1182_blood-2014-02-556993/4/m_393f1.jpeg?Expires=1769307317&Signature=Hgdd-zie5CXK1q0TLP-bPcd7t6dQ4Zujuv4jbVa-bWjFP7YIjTPvrIXPIFj6UyfPMKtszfld2dlnxnVK9lffl7olJlR~DdF6sHbkTXHqN~Jrb-DQ~2nkE4hhITX8swigptskEZidtReIWqNvbDICcWWq39Sx2xKBsv3CH~L7dgJNmXKWhdLmjb-~gaNcBWRC-QDkbPC0ToTPbyOLS40QjO9aGLCnfzn~xhyqtid4Lgiq7EFzlUeLZaYPCcntXEDl03ttDEsYIaASg1HJvvnaQQPAybTeoYofaBa~ygfxk2a6VlC7CUtwWtSDK-l-ehcY2XLPUwi8gS9D0tcw6hoItg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 1. Conditional depletion of Smad1 and Smad5 reveals differing contributions of each to primitive hematopoiesis. Cre/loxP-mediated homologous recombination was used to target insertion of constructs containing shRNA hairpins corresponding to sequences within the MH1 domain of Smad1 alone (miSMAD1), Smad5 alone (miSMAD5), or both Smad5 and Smad1 (miSMAD51). Each hairpin construct is targeted to the identical site downstream of an operator/promoter, which allows tet-on activation via dox treatment (miSMAD1 was previously characterized33). (A) Representative western blotting analysis of SMAD5 and SMAD1 protein expression in lysates derived from EBs after 24 hours of dox treatment (DOX, compared with uninduced control). (B) Quantitative densitometric analysis of samples induced with 2 μg/mL dox normalized to control β-actin levels from at least 3 separate experiments to derive mean ± SEM. (C-E) Embryoid bodies derived from parental AinV18 cells, miSMAD5, miSMAD51, or miSMAD1 ESCs were induced with dox on day 2 (dox d2) or day 4 (dox d4) of EB differentiation and evaluated by hematopoietic colony assays to measure primitive erythroid (EryP [C]) and myeloid (macrophages, MacP [D]; megakaryocytes, MegaP [E]) potential. Colony assays were each performed at least 3 separate times, with samples counted in triplicate, and values are reported as normalized to untreated controls. Statistical significance compared with no dox controls, with *P < .05 and **P < .01, respectively. CTL, control; dox, doxycycline.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/3/10.1182_blood-2014-02-556993/4/m_393f1.jpeg?Expires=1769307318&Signature=d0k0er7IOtjCOVl8JYaMCHu0kYzgRpWXZxPCTSVw2JQILdxhrm6M3gESUkgmU4lbkliLJrsDF6aQeQyiBvrMf6uGZLwk3sXswQBgWvkknas5U3p-a5S6EHb1cAti6KDos4~lLpQn4FK0odgVj0WUKqLDa0C-ndzD-gcF9DxZ~957wtA0SK5wVEclH7KylDGk4pqUyFVYxAR6ve8gQgAqj1FZoQhdxyWIeAvxcn9RI7zOzOtJy3Ia7ZZXTogc6x8tN4AnSzL0Q-q-ZYLDndK13aAtl948QLgwiAJezw0yWqxfQclQWR7is7IhEG3sk970wodSycPPkkENp0~iudAsVA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)