Abstract
Mouse models that recapitulate human malignancy are valuable tools for the elucidation of the underlying pathogenetic mechanisms and for preclinical studies. Several genetically engineered mouse models have been generated, either mimicking genetic aberrations or deregulated gene expression in chronic lymphocytic leukemia (CLL). The usefulness of such models in the study of the human disease may potentially be hampered by species-specific biological differences in the target cell of the oncogenic transformation. Specifically, do the genetic lesions or the deregulated expression of leukemia-associated genes faithfully recapitulate the spectrum of lymphoproliferations in humans? Do the CLL-like lymphoproliferations in the mouse have the phenotypic, histological, genetic, and clinical features of the human disease? Here we compare the various CLL mouse models with regard to disease phenotype, penetrance, and severity. We discuss similarities and differences of the murine lymphoproliferations compared with human CLL. We propose that the Eμ-TCL1 transgenic and 13q14-deletion models that have been comprehensively studied at the levels of leukemia phenotype, antigen-receptor repertoire, and disease course show close resemblance to the human disease. We conclude that modeling CLL-associated genetic dysregulations in mice can provide important insights into the molecular mechanisms of disease pathogenesis and generate valuable tools for the development of novel therapies.
Introduction
Chronic lymphocytic leukemia (CLL) originates from the clonal expansion of mature B cells, which show features of antigenic stimulation and express the CD5 cell surface antigen. CLL is a complex disease in which genetic abnormalities cooperate with microenvironmental factors in the malignant transformation of the tumor-cell precursor and in leukemia progression.1 Compared with most other subtypes of non-Hodgkin lymphoma (NHL), CLL shows a lower frequency of genetic mutations per case and a different spectrum of genetic aberrations, which mostly comprise chromosomal deletions (13q14, ATM, and TP53) or amplifications (trisomy of chromosome 12).2,3 More recently, next-generation sequencing (NGS) analyses have identified several novel recurrent mutations in CLL, including those that target the NOTCH1, MYD88, SF3B1, and BIRC3 genes.4-11 A number of genes are overexpressed in CLL tumor cells compared with normal lymphocytes, presumably as a direct consequence of the genetic aberrations (eg, BCL2 and MCL1 due to deletion of mir-15a/16-112 ) or through as yet unknown mechanisms (eg, ROR113,14 or TCL115 ). Finally, genome-wide association studies have identified several susceptibility loci for familial CLL,16 including a single nucleotide polymorphism in the IRF4 gene, a known regulator of B-cell developmental processes.17
Genetically engineered mouse models of CLL
Several mouse models mimicking genetic lesions found in CLL (13q14 deletion), transgenic for genes that are overexpressed in the disease (including TCL1, APRIL, BCL2 × traf2dn, ROR1), or driven by ectopic oncogene expression (IgH.T and IgH.TEμ) have been generated (Table 1).
Mouse models mimicking the spectrum of deletions of chromosomal region 13q14
Deletion of 13q14 is the most frequent genetic lesion in CLL that occurs in >50% of cases and is clinically associated with an indolent disease with no or delayed need for therapy.3,18 The deletion is detected also in monoclonal B-cell lymphocytosis (MBL),19 an expansion of CD5+ B lymphocytes in the peripheral blood (PB) of otherwise healthy individuals19,20 that is thought to precede CLL,21 and is present at a lower frequency in other NHL subtypes.22
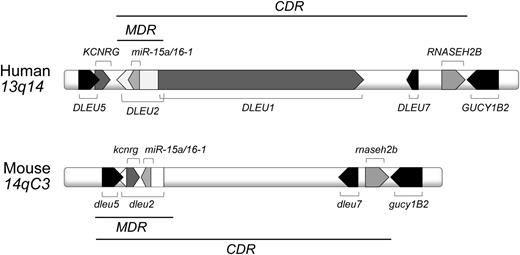
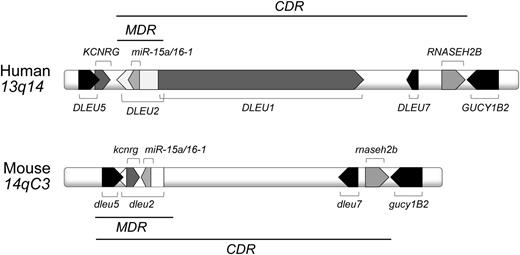
The 13q14 region corresponds to the murine 14qC3 locus and encodes several genes that are highly conserved among human and mouse (Figure 1). A minimal deleted region (MDR) includes the first exon of the DLEU1 sterile RNA, the deleted in leukemia (DLEU) 2 gene, encoding a sterile transcript and the miR-15a/16-1 cluster, located in an intron of DLEU2.23-25 A larger 13q14 deletion is found in a sizable number of CLL cases and is hence named the common deleted region (CDR). The functional dissection of the 13q14 tumor suppressor locus by using transgenic mouse models demonstrated that the size of 13q14 deletions influences the phenotype of the developing lymphoproliferations and the severity of disease (Table 1),26,27 suggesting a tumor suppressor function for multiple genetic elements encoded in the deleted region. Specifically, the results provided evidence for a causal role of the microRNAs mir-15a/16-1 in CLL pathogenesis and demonstrated that the additional deletions of dleu2 and/or dleu5 caused a higher disease penetrance and a more severe disease course.26 In both models, CD5-negative NHLs developed along with CD5-positive lymphoproliferations that encompassed MBL and CLL. Importantly, CDR-deleted mice were characterized by a different spectrum of lymphoproliferations compared with MDR and mir-15a/16-1–deleted mice, as they mostly developed CLL (rarely MBL or NHLs), and a more aggressive disease course.27 A commonality among all 13q14 mouse models was the expression of unmutated stereotypic antigen receptors,26,27 indicating a role of antigen in the expansion of the CLL clone.
Human 13q14 and murine 14qC3 locus. Indicated are the MDR and the CDR.
Mouse models mimicking the deregulated expression of genes in human CLL
Several mouse models have been generated that are transgenic for genes overexpressed in the disease (Table 1). The first transgenic mouse that developed a CLL-like disease and that since has been widely used in the study of CLL pathogenesis and therapy is the Eμ-TCL1 transgenic mouse.28,29 The TCL1 gene is expressed in the vast majority of human CLL cases.15 Strong TCL1 expression is associated with markers of poor prognosis, including unmutated IGHV genes.15 Eμ-TCL1 mice developed a clonal expansion of CD5+IgM+ B cells initially in the peritoneal cavity and on progression in the PB, spleen, and bone marrow,28,29 eventually giving rise to a monoclonal B-cell population in virtually 100% of mice (Table 1).28 The B-cell receptors (BCRs) expressed by lymphoproliferations in Eμ-TCL1 mice harbored unmutated IGHV gene rearrangements and exhibited stereotypy in IGHV, IGKV, and IGLV gene rearrangements.30
Commonalities and disparities among CLL mouse models
In all mouse models, the lymphoproliferations develop late in life and resemble the indolent disease course of CLL. Also, the target cell of the malignant transformation to CLL seems to be the peritoneal B1a cell, as suggested by the expression of CD5 and of unmutated IGHV genes, high levels of IgM, and low levels of IgD and CD23, at least in the models where these parameters were analyzed. Noticeably, whereas virtually all Eμ-TCL1 mice developed CLL, a sizable fraction of B-cell lymphoproliferations in the 13q14 deletion models represented CD5-negative NHLs. The latter observation genuinely reflects the heterogeneous spectrum of B-cell malignancies with the 13q14 deletion occurring in humans.
The most notable difference among the CLL mouse models is the penetrance of the phenotype, which is highest in the Eμ-TCL1 mice (∼100%), intermediate in the 13q14-MDR and CDR deletion models and the APRIL transgenic mice (40-50%), and lowest in the mir-15/16-1 deletion model (∼25%) and the ROR1 transgenic mice (5%) (Table 2). The different tumor incidence among the CLL mouse models suggests that the various genetic aberrations predispose to, but are not sufficient per se, for the malignant transformation of B cells, a situation similar to that encountered by 13q deletions in low-count MBL, which virtually never progresses into frank leukemia.20 Thus, additional alterations are required to induce leukemia. The heterogeneity of the lymphoproliferation phenotypes observed in mir-15a/16-1−/− and MDR−/− mice (CLL, MBL, and CD5– NHL) may reflect the specific nature of the secondary hits and/or the different B-cell differentiation stages in which the malignant transformation occurs. In particular, the observation that CDR-deleted mice predominantly develop a CLL-like disease27 suggests the existence of tumor suppressor genes in the h13q14/m14qC3 locus in addition to the DLEU2/miR-15a/16-1 cluster. The almost complete disease penetrance in the Eμ-TCL1 model is likely due to a strong oncogenic function of the hTCL1 gene, which affects multiple pathways, including coactivation of the serine/threonine kinase protein kinase B/v-AKT murine thymoma viral oncogene (AKT),38,39 activation of the nuclear factor κB pathway,40 and inhibition of the activity of DNA methyltransferase (DNMT)3A and DNMT3B.41 In the APRIL transgenic model, the sustained microenvironmental stimulation likely facilitates the expansion of B1a cells, which transform in 40% of cases.32 Similarly, prosurvival signals delivered through the ROR1 receptor may favor the expansion of a preleukemic B-cell population that gives rise to a CLL-like disease in 5% of animals.34 Conversely, an increased proliferative capacity seems to account for leukemia development in 20% of Eμ-mir-29 transgenic mice.35 Overexpression of BCL2 synergizes with a dominant negative form of the adaptor protein tumor necrosis factor (TNF) receptor-associated factor 2 (traf2dn) in the malignant transformation of B cells, resulting in the development of a CLL-like disease in ∼80% of BCL2 × traf2dn transgenic mice, most likely due to resistance to apoptosis and altered expression of adhesion molecules.42 Apoptosis resistance has also been suggested to account for leukemia development in the Vh11 × irf4−/− model,36 although a recent study suggests that the possible leukemogenic function of reduced IRF4 expression may be the alteration of the migration properties of B cells,43 presumably through up-regulation of NOTCH2 activity in IRF4-deficient cells.43 Finally, expression of the simian virus 40 (SV40) T antigen in B cells has been associated with the induction of genomic instability and deregulation of genes involved in proliferation, DNA repair, and apoptosis.37 The almost complete disease penetrance in the latter 2 mouse models is likely to be due to a skewed IGHV gene repertoire (the Vh11 × irf4−/−) and the ectopic expression of a potent oncogene (SV40 T antigen), respectively, thus introducing exogenous factors that accelerate B-cell transformation. Therefore, among the models that mimic genetic dysregulations found in human CLL, overexpression of the hTCL1 gene in mouse B cells accounts for the highest disease penetrance.
There is also the perception that the disease developing in Eμ-TCL1 mice is more aggressive compared with the one in the 13q14 deletion models. Indeed, based on the occurrence of unmutated BCRs in Eμ-TCL1 leukemic mice that resemble those in human CLL and are structurally similar to autoantibodies or antibodies recognizing microbial antigens, it was proposed that CLL developing in those mice correspond to the aggressive, treatment-resistant subtype in humans. However, the IGHV genes of the leukemic clones developing in Eμ-TCL1 and 13q14 deletion models were surprisingly similar. A sizable number of rearrangements obtained from leukemias in both of these models could be assigned to 5 stereotype sets defined by Chiorazzi et al.30 Even more intriguing, CLLs with identical IGHV-IGHD-IGHJ junctions were found among the Eμ-TCL1 and 13q14-deleted mouse models.27 These observations suggest that the same antigens are involved in the transformation and progression of B-cell clones developing in Eμ-TCL1 transgenic and 13q14-deleted animals. Moreover, these models show a similar disease phenotype that is characterized by the infiltration of the leukemic cells in lymphoid and nonlymphoid organs. Together with evidence that supports a common cell of origin (see below), this argues against a difference in disease aggressiveness among the Eμ-TCL1 transgenic and 13q14-deleted models. The shorter latency of the appearance of a B1a cell expansion (4-5 months in Eμ-TCL1 mice vs 9-12 months in the 13q14 deletion models) and higher penetrance of the phenotype suggest that the hTCL1 oncogene causes the expansion of a larger pool of CLL precursor cells that are targets for additional transforming events in Eμ-TCL1 compared with 13q14-deleted mice. Interestingly, relative to the mir-15a/16-1−/− and MDR−/− models, there is no obvious MBL stage in Eμ-TCL1 transgenic and CDR-deleted mice that precedes CLL development, which may be indicative of a faster disease progression. However, it seems that after a malignant clone has appeared, Eμ-TCL1 and 13q14-deleted animals show the same disease course.
Commonalities among CLL developing in mouse models and humans
The usefulness of CLL mouse models for the study of the human disease depends on the grade of relatedness of the disease characteristics between animal models and patients. There are 2 issues to consider. First, with respect to the involvement of the genetic lesions or the deregulated expression of leukemia-associated genes, do the models faithfully recapitulate the spectrum of lymphoproliferations observed in the human disease? Second, do the CLL-like lymphoproliferations in the mouse have the phenotypic, histological, genetic, and clinical features of the disease developing in humans? Evidently, a major factor in this is the phenotypic and functional relatedness of the cells targeted by the oncogenic transformation in humans and mice. In the following, we will discuss the commonalities among CLL in mice and humans with respect to disease origin, repertoire of antigen receptors, and phenotypic and epigenetic characteristics of the tumor cells.
Disease origin
Possibly all CD5-positive lymphoproliferations developing in CLL mouse models derive from the transformation of peritoneal, self-replenishing CD5+ B1a cells.44 Clonal populations of B1a cells develop spontaneously within the peritoneal cavity of aging mice and eventually disseminate to and expand in other lymphoid organs.45,46 This suggests that over time, B1a cells become susceptible to malignant transformation as the results of genetic abnormalities that accumulate during their self-replenishment, eventually giving rise to a CLL-like disease. Protumorigenic forces such as deletion of 13q14, overexpression of TCL1, or high serum levels of APRIL that confer a proliferation and/or survival advantage may accelerate the disease onset, increase the penetrance of the phenotype, and impact disease course and severity (Table 2).
What is the relationship between the CLL precursor cells in the human and mouse? The longstanding discussion about the putative cell of origin of CLL in humans has recently been revisited by Chiorazzi and Ferrarini,47 who provide conceivable arguments for the not mutually exclusive possibilities that the oncogenic transformation may occur in a marginal zone B cell, a transitional B cell, or a human B1-like cell,47 and independent evidence suggests that CLL cases with unmutated IGHV genes may originate from transformed naïve B cells.48 The notion of a B1a-like cell of origin of CLL recently gained traction through the identification of a previously unrecognized CD5-expressing B-cell population in the adult.49 These cells comprise only about 1% of PB B cells, but their gene expression profile is closely related to that of CLL cells.49 One notable aspect of this new hypothesis is that it would be consistent with the fact that the CD5 antigen is invariably expressed on human and murine CLL cases. However, because the function of these newly identified CD5+ B cells is unknown, it is presently impossible to draw a functional relationship between these cells and murine B1a cells.
Stereotypic antigen receptors
A recent study provided a fascinating example that the expansion of the CLL clone can be driven by antigen-independent cell autonomous signaling.50 Nevertheless, there is clear evidence for the role of antigen-BCR stimulation via autoantigens or external antigens in CLL pathogenesis.51-54 A characteristic of CLL is the expression of stereotypic antigen receptors among unrelated CLL cases that show highly similar heavy-chain complementarity-determining (HCDR3) regions,55 presumably as the result of strong selection forces because the normal HCDR3 repertoire is extremely diverse due to IGHV-IGHD-IGHJ combinatorial and junctional diversity. Sequence analyses of antigen-receptor rearrangements from Eμ-TCL1 transgenic and 13q14 deletion models demonstrated a skewed IGHV-IGHD-IGHJ rearrangement repertoire in the leukemic clones derived from these mice,26,27,30 which is reflected by the expression of highly similar HCDR3 regions that are reminiscent of human stereotypic antigen receptors. Specifically, 44% of the sequences from the Eμ-TCL1, miR-15a/16-1, MDR, and CDR cohorts could be assigned to 8 sets of stereotypic HCDR3 regions, which are defined by ≥80% homology at the protein level.27 This finding clearly suggests that, similarly to the human disease, discrete antigens or classes of structurally related epitopes select and activate the leukemic clones in the corresponding mouse models and provides additional molecular evidence for the similarity of the disease developing in humans and the CLL mouse models.
Phenotypic and epigenetic characteristics
The ZAP70 tyrosine kinase is recruited to the cell surface of CLL cells on stimulation through the BCR and promotes activation of the downstream signaling pathway.56,57 In addition, ZAP70+ CLL cells display increased migration properties.58 ZAP70 expression is up-regulated in splenic B cells of Eμ-TCL1 mice59 already in the early stage of the disease (ref. 59 and G.S., M.T.S.B, and P.G., unpublished data, 2008), suggesting an involvement of the kinase during the clonal expansion. Deleting zap70 in the B-cell compartment of Eμ-TCL1 mice may provide new insights into the role of ZAP70 in the dynamics of disease onset, progression, and dissemination.
Recent studies suggest that CLL cells have the functional capacity to express and secrete interleukin (IL)-10,60 a property that characterizes a subset of B cells with immune regulatory functions (B-10 cells).61 Based on the observation in Eμ-TCL1 mice that the expansion of IL-10–competent malignant B cells precedes the development of overt leukemia, IL-10 production was suggested to participate in the immunoregulatory capacity of the malignant cells and therefore influence disease progression, outcome, and response to treatment,60 possibly through suppression of the antitumor response.
As observed in CLL patients, leukemic Eμ-TCL1 mice display multiple T-cell alterations, including the shift from a naïve to a memory T-cell subtype62 and defective signal transduction at the immune synapse.63 Also, similar to human CLL,64 leukemic mice present with an increased number of T cells in the PB.62 Interestingly, transfer of leukemic B cells into disease-free Eμ-TCL1 mice caused the development of T-cell dysfunctions that allowed the malignant clone to escape from the host immune system and expand in the recipient mice.63 Together, these findings suggest that the Eμ-TCL1 model represents a suitable tool to study the interplay between CLL cells and T cells in disease development and progression.
CLL cells in humans show multiple epigenetic alterations that have been detected also in the Eμ-TCL1 model.65 Aberrant methylation of promoter sequences was observed in B cells of Eμ-TCL1 mice starting at 3 months of age, and the level of DNA methylation increased with time, suggesting a causative role in the development and progression of CLL.66 In addition, splenic B cells from Eμ-TCL1 mice showed an elevated number of hypomethylated regions compared with wild-type B cells.41
CLL mouse models in the study of disease pathogenesis and therapy
Eμ-TCL1 transgenic mouse model as a tool to investigate pathogenic mechanisms
Due to the similarity of the disease characteristics of CLL in humans and mice and the complete penetrance of the model, Eμ-TCL1 mice have been extensively used in the dissection of the pathogenic mechanisms leading to CLL. Several transgenic and knockout mouse models have been crossed with Eμ-TCL1 mice to elucidate the functional role of specific molecules in the onset and progression of CLL in vivo (Figure 2; supplemental Table 1), providing important new insights into the pathogenic role of those genes in the dysregulation of signaling, proliferation, and apoptosis, in mediating altered trafficking and homing, and in the aberrant cross-talk with the microenvironment.
Study of novel pathogenic mechanisms of CLL in the TCL1-driven leukemia model. Overexpression (transgenic [tg]) or deficiency of different molecules in the Eμ-TCL1 transgenic mouse model variably affects the disease phenotype (BM, bone marrow; TAM, tumor-associated macrophages). The following mouse models have been crossed with Eμ-TCL1 mice: xid,91 pkcβ−/− (or pkcβ+/−),70 xbp1fl/flCD19-Cre,73 tir8−/−,72 dnrag1-tg,71 hs1−/−,79 rhoh−/−,59,80 p53−/−,75 id4−/−,76 miR29a/b-tg,35 fzd6−/−,89 cd44−/−,87 mif−/−,86 ROR1-tg,34 APRIL-tg,85 and baff-tg.84
Study of novel pathogenic mechanisms of CLL in the TCL1-driven leukemia model. Overexpression (transgenic [tg]) or deficiency of different molecules in the Eμ-TCL1 transgenic mouse model variably affects the disease phenotype (BM, bone marrow; TAM, tumor-associated macrophages). The following mouse models have been crossed with Eμ-TCL1 mice: xid,91 pkcβ−/− (or pkcβ+/−),70 xbp1fl/flCD19-Cre,73 tir8−/−,72 dnrag1-tg,71 hs1−/−,79 rhoh−/−,59,80 p53−/−,75 id4−/−,76 miR29a/b-tg,35 fzd6−/−,89 cd44−/−,87 mif−/−,86 ROR1-tg,34 APRIL-tg,85 and baff-tg.84
As predicted from the study of CLL in humans,67,68 the establishment of suitable Eμ-TCL1 compound mouse models could validate the critical role of BCR signaling in CLL pathogenesis,69,70 provided evidence for the importance of continuous autoantigenic stimulation in CLL development,71 and suggested a proleukemogenic function of Toll-like receptor signaling.72 Interestingly, a recent study found that B cell-specific inhibition of the endoplasmic reticulum stress response, which is coupled to defective spleen tyrosine kinase and Bruton agammaglobulinemia tyrosine kinase activation on BCR stimulation, reduces leukemia development in Eμ-TCL1 mice.73
As expected from the known tumor suppressor function of TP53, genetic ablation of tp53 in mice, thus mimicking 17p deletion,3 the most deleterious genetic abnormalities in CLL,74 accelerated leukemia onset and progression in the Eμ-TCL1 model.75 Similar observations were made for a putative CLL tumor suppressor gene: inhibitor of DNA binding protein 4 (id4).76
The notion that aberrant cytoskeletal organization and cell trafficking contribute to CLL pathogenesis potentially through a disrupted interaction of the malignant cells with the bone marrow microenvironment77,78 was experimentally confirmed in vivo in hematopoietic cell-specific Lyn substrate (hs1)−/−Eμ-TCL1 mice.79 In addition, another Eμ-TCL1 compound mouse model provided evidence that cellular motility plays an important role in bone marrow infiltration of CLL cells.59,80
In CLL patients, endothelial, nurse-like, and antigen-presenting cells release the TNF family members B-cell activating factor (BAFF) and APRIL, which activate the transformed B cells.81-83 In accordance, ectopic expression of the corresponding transgenes in Eμ-TCL1 mice revealed that BAFF and APRIL protect leukemic cells from apoptosis in vivo, resulting in a rapid expansion of the malignant clone.84,85 The importance of microenvironmental interactions in the maintenance of the CLL clone could further be demonstrated by 2 independent Eμ-TCL1 compound mouse models (mif−/−Eμ-TCL1 and cd44−/−Eμ-TCL1 mice).86,87 Moreover, the potential role of the tumor microenvironment in signaling to malignant B cells through wingless-type MMTV integration site family (WNT) receptors88 was addressed by genetically ablating the WNT receptor frizzled 6 (fzd6) in Eμ-TCL1 mice. The results suggested that dysregulated FZD6 expression modulates the disease course as it delayed but did not abrogate tumor growth.89
In summary, Eμ-TCL1 compound mouse models have proved extremely useful in determining the pathogenic functions of CLL-associated genes in an in vivo context. It will be interesting to explore the leukemogenic role of the newly identified oncogenic mutations in the NOTCH1, MYD88, SF3B1, and BIRC3 genes4-11 by modeling these mutations on the background of Eμ-TCL1, as well as other CLL mouse models.
Eμ-TCL1 transgenic mouse model as a preclinical model for novel therapeutics
Mouse models represent a useful tool to evaluate efficacy, pitfalls, and potential side effects of novel therapeutic strategies. The Eμ-TCL1 mouse has been validated as a suitable preclinical model by demonstrating that the response of leukemic mice to fludarabine, a cytotoxic drug used in first-line treatment of CLL, resembles that observed in human patients.29 The disadvantage of the long disease latency was overcome by transferring splenic leukocytes or B cells from leukemic mice into syngeneic or immunodeficient recipient mice.34,84,87,90-99 Generally, transplanted animals developed a frank leukemia within a few weeks or months. Together with xenograft models of CLL,100 such transfer experiments represent reproducible systems for the elucidation of the efficacy and mechanism of action of novel therapies. As such, preclinical studies with Eμ-TCL1 mice were instrumental in understanding the pathogenic relevance of molecular targets in disease biology (supplemental Table 2), as summarized in the following.
Inhibition of signaling
Based on the known function of the TCL1 oncogene as an AKT coactivator,38,39 the dependence of TCL1-driven leukemia on AKT signaling has been investigated by inhibiting the mTOR effector with rapamycin in a transfer model.90 The study demonstrated a critical role of the AKT pathway in the disease onset rather than progression.
The fact that CLL cells are chronically stimulated and in lymphoid tissues show a gene expression signature of lymphocytes activated through the BCR101 provided a rationale for the development of strategies based on BCR signaling inhibitors. In clinical practice, the Bruton agammaglobulinemia tyrosine kinase inhibitor ibrutinib (PCI-32765), the spleen tyrosine kinase inhibitor fostamatinib, and the phosphatidylinositol 3-kinase-δ inhibitor idelalisib (GS-1101) caused rapid lymph node shrinkage and transient lymphocytosis in patients102,103 by impairing the retention of CLL cells in the tissue microenvironments.104 Importantly, a transient increase in PB B-cell counts has also been reported early after treatment of mice transplanted with Eμ-TCL1–derived leukemias with fostamatinib and ibrutinib.92,93
Inhibition of cytoskeletal functions and cell trafficking
The findings that BCR inhibitors cause rapid tissue mobilization of CLL cells established a new paradigm in the CLL field, which identifies the disruption of the homing properties of leukemic cells as a therapeutic approach. This notion is supported by results from HS1 or RHOH-deficient Eμ-TCL1 compound mouse models (supplemental Table 1),79,80 by recent phase 3 studies,105 and by the Food and Drug Administration approval of ibrutinib for the treatment of CLL. In line with these assertions, interfering with the proper cytoskeleton function of CLL cells via the dual v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (SRC) kinase/BCR-Abelson murine leukemia viral oncogene homolog 1 (ABL) inhibitor dasatinib may be a conceivable therapeutic option for the subset of patients with an active v-yes-1 yamaguchi sarcoma viral related oncogene homolog (LYN)/HS1 axis.98 Indeed, inhibition of LYN/HS1 signaling impaired leukemia progression and lymphoid organ infiltration in a transfer model of TCL1-driven leukemia. Moreover, an indirect effect on leukemic cell trafficking may contribute to the efficacy of lenalidomide, an immunomodulatory agent that is currently under investigation as salvage therapy for patients with relapsed or refractory CLL106 and as a consolidation strategy in progressive cases.107 Lenalidomide treatment reduced the chemokine response of CLL cells in vitro and down-modulated the expression of RHOH,80 which, in the Eμ-TCL1 model, is indispensable for the homing of malignant B cells to the bone marrow.80
Inhibition of tumor-microenvironmental interactions
Several novel therapeutics target the cross-talk between the malignant cells and the tumor microenvironment through neutralization of protumorigenic factors, inhibition of surface receptors, or nuclear transport. Treatment with the anti-CD44 antibody IM7 that exerts a proapoptotic function prevented the accumulation of CD44-expressing leukemic B cells in the PB of an adoptive transfer model of TCL1-driven leukemia early after injection.87 Similarly, administration of the anti-ROR1 monoclonal antibody D10 inhibited the engraftment of leukemic cells from ROR1 × Eμ-TCL1 transgenic mice into ROR1 transgenic recipients, which was accompanied by a strong reduction of PB and splenic B-cell counts.34 A reduction in the number of circulating malignant B cells was also observed on neutralizing the prosurvival effect of BAFF with CD268-Fc and CD269-Fc decoy receptors in a model obtained by intraperitoneal injection of CD5+ leukemic B cells from baff × Eμ-TCL1 transgenic mice into baff transgenic recipients.84 Finally, marked antileukemic effects have recently been reported for the inhibitor of nuclear export KPT-185 and the antibody drug conjugate IMGN529.95,97 KPT-185 acts by blocking the activity of the nuclear exporter chromosome region maintenance 1 protein, or exportin1 (CRM1/XPO1), which is a downstream mediator of microenvironmental signals sustaining CLL cell survival and growth.97 Administration of the small molecule to SCID mice engrafted with leukemic B cells from Eμ-TCL1 mice improved survival of those mice. A benefit in terms of overall survival has also been observed in CD37 transgenic mice engrafted with CD37 × Eμ-TCL1 leukemia on treatment with IMGN529, which consists of the cytotoxic anti-CD37 antibody and the antimicrotubule agent DM1 that exerts an antiproliferative effect.95
Forced induction of apoptosis
A major pathogenic mechanism in CLL development constitutes the resistance of the tumor cells to apoptosis. Therefore, therapeutics that target the antiapoptotic pathway represent valuable strategies for CLL treatment. Preclinical studies of actinomycin D108 and silvestrol (a plant-derived cyclopenta[b]benzofuran)109 that, respectively, inhibit transcription and translation, and of 17-dimethylaminoethylamino-17-demethoxygeldanamycin (17-DMAG) (a derivative of the heat shock protein 90 inhibitor geldanamycin)94 showed promising results and were indeed associated with a reduction in the levels of antiapoptotic proteins. The studies may provide a rationale for the treatment of CLL cases carrying a deletion of the 17p locus (that are resistant to standard chemotherapy including fludarabine)110 with actinomycin D or silvestrol, which can induce apoptosis in a P53-independent fashion.108,109
Epigenome targeting
Finally, CLL cells display epigenetic alterations, and evidence suggests that overexpression of histone deacetylase enzymes contributes to CLL pathogenesis.65,111 The histone deacetylase inhibitor MGCD0103 induced apoptosis112 and autophagy suppression113 in CLL cells in vitro and showed antitumor activity in a phase 2 clinical trial.114 Moreover, the class I and II deacetylase (DAC) inhibitor, AR-42, prolonged the overall survival of mice transplanted with TCL1-driven leukemia.99 Class I and II inhibitors such as AR-42 that target both histone and nonhistone substrates may be promising therapeutics for a multitarget therapy.
Conclusions and outlook
Several genetically engineered mouse models of CLL have been generated that mimic the human disease. The 13q14 deletion models provided the first in vivo evidence of the tumor-suppressor function of a CLL-associated genetic lesion. As numerous studies revealed similarities between CLL developing in humans and Eμ-TCL1 mice, this transgenic model has become a suitable tool in the investigation of pathogenic mechanisms of CLL and represents a convenient preclinical model, particularly due to its complete disease penetrance.
Based on the stereotypic antigen receptors in the Eμ-TCL1 mice that recall those in some aggressive forms of human CLL, and the association of the 13q14-deletion in humans with CLL bearing somatically mutated IGHV genes that have a more favorable disease course, it has been suggested that the corresponding mouse models mimic the aggressive and indolent forms of CLL, respectively. However, this view may have to be revisited due to the circumstance that the CD5+ lymphoproliferations developing in the Eμ-TCL1 transgenic and 13q14 deletion models are similar regarding their phenotype and their expression of unmutated IGHV genes and of stereotypic antigen receptors, indicating that the target cell of the oncogenic transformation may be the same in the 2 models. Two implications ensue from this: first, in lieu of knowledge on the precise cell of origin of CLL in humans, it is impossible to confidently assign the CLL developing in the mouse models to a particular human CLL subtype. Second, the virtual absence of somatically mutated IGHV genes in CLLs developing in the mouse models may suggest that the normal cellular counterpart(s) of CLL differ among mice and humans, eg, that a somatically mutated CD5+ subset may simply not exist in the mouse. In support of the latter assumption is the observation that, similar to humans, CD5-negative NHL with somatically mutated IGHV genes do develop in 13q14 deletion models.
Regardless of these unresolved issues, genetically engineered CLL mouse models, together with xenograft models, have emerged as valuable tools for the first-line testing of new therapies. A downside of the transgenic CLL models in their use as preclinical models is the long latency until disease develops. The presence of stereotypic receptors and active BCR signaling in the leukemic cells of those mice may provide a rationale for the development of a CLL mouse model with potentially shorter latency by combining an antigen or BCR-driven mechanism with the commonly known, or newly identified, CLL-associated genetic aberrations.
The online version of this article contains a data supplement.
Acknowledgments
The authors thank Riccardo Dalla-Favera and Federico Caligaris-Cappio for discussions and support.
This project was supported by Associazione Italiana per la Ricerca sul Cancro (Investigator Grant and Special Program Molecular Clinical Oncology–5 per mille #9965) and Ricerca Finalizzata 2010–Ministero della Salute, Roma, and by a grant from the CLL Global Research Foundation.
Authorship
Contribution: G.S., M.T.S.B., P.G., and U.K. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for G.S. is Department of Experimental, Diagnostic and Specialty Medicine, University of Bologna, Bologna, Italy.
Correspondence: Ulf Klein, Herbert Irving Comprehensive Cancer Center, Columbia University, 1130 St. Nicholas Ave, New York, NY 10032; e-mail: uk30@columbia.edu; or Paolo Ghia, Università Vita-Salute San Raffaele, Via Olgettina 58, 20132 Milan, Italy; e-mail: ghia.paolo@hsr.it.

![Figure 2. Study of novel pathogenic mechanisms of CLL in the TCL1-driven leukemia model. Overexpression (transgenic [tg]) or deficiency of different molecules in the Eμ-TCL1 transgenic mouse model variably affects the disease phenotype (BM, bone marrow; TAM, tumor-associated macrophages). The following mouse models have been crossed with Eμ-TCL1 mice: xid,91 pkcβ−/− (or pkcβ+/−),70 xbp1fl/flCD19-Cre,73 tir8−/−,72 dnrag1-tg,71 hs1−/−,79 rhoh−/−,59,80 p53−/−,75 id4−/−,76 miR29a/b-tg,35 fzd6−/−,89 cd44−/−,87 mif−/−,86 ROR1-tg,34 APRIL-tg,85 and baff-tg.84](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/7/10.1182_blood-2014-05-577122/5/m_1010f2.jpeg?Expires=1765883738&Signature=n6iyO781iAaBrN00zNItawTf1xzSurhdmNNNIV7PxT0T5N0kM8KCtDk5Oam08Fm~M6NtrCQrcyn3JZZFlmFgvAFW4BPbE0mewQYzuSKawVCfEaj4A2vGv52vAk~kfdeBNVyVbXb2d9Ee1GKIOPuIrUmKUiV57QlO~DtHuIlMjquLZPsVV6ojKoQOvJBeSxNqX5D-HCWrxkSoi1bP9DtvlyvPm5tl2rAM3f8peXFDtWJh3KhEiO6s8eCNSOa9PRlKNuXvxsz0Ok3nNZbJL2d4LPj6R46hrQDUHEKAhJ73hK6bpc5Ha1QRsmITyWkMpv7JP1sso2qSp-HWGOzyCpRMGA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 2. Study of novel pathogenic mechanisms of CLL in the TCL1-driven leukemia model. Overexpression (transgenic [tg]) or deficiency of different molecules in the Eμ-TCL1 transgenic mouse model variably affects the disease phenotype (BM, bone marrow; TAM, tumor-associated macrophages). The following mouse models have been crossed with Eμ-TCL1 mice: xid,91 pkcβ−/− (or pkcβ+/−),70 xbp1fl/flCD19-Cre,73 tir8−/−,72 dnrag1-tg,71 hs1−/−,79 rhoh−/−,59,80 p53−/−,75 id4−/−,76 miR29a/b-tg,35 fzd6−/−,89 cd44−/−,87 mif−/−,86 ROR1-tg,34 APRIL-tg,85 and baff-tg.84](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/7/10.1182_blood-2014-05-577122/5/m_1010f2.jpeg?Expires=1765938504&Signature=GQ8c6DzU5zWOuXXYqEojDbiUHiu2OHVCVs5eDJFDPPWOryR1X1uOYXwIY-ko6TtuJgJlrAaga66afjisog2RLGrNqdZeb3Id~9-Ufoc90PC6rKdEuDrQN0vBPTXbq~awu8cmvbnJFGz8qxsy3DscT~OW~fy-XaS-HcFtdkpWfoej3OgfkvNCFJ~F8a9gGMK0~YpMyjPQxfyz6kYSS7BOyzHKtS04zzh6QCTEShpCvKb2Bh6Av9k6tfZIV8DP8LCcxivMO2FICaepkATbVykJmdkmZ~NyznplX5swkCLKr8~GvxleTdOKUiKQB3gfvpmZqYvVygBHEua-gWsxinhjxQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)