Key Points
A comprehensive analysis of the ADAMTS13 Cys-rich domain identifies a novel functional interaction between ADAMTS13 and VWF.
Abstract
ADAMTS13 proteolytically regulates the platelet-tethering function of von Willebrand factor (VWF). ADAMTS13 function is dependent upon multiple exosites that specifically bind the unraveled VWF A2 domain and enable proteolysis. We carried out a comprehensive functional analysis of the ADAMTS13 cysteine-rich (Cys-rich) domain using engineered glycans, sequence swaps, and single point mutations in this domain. Mutagenesis of Cys-rich domain–charged residues had no major effect on ADAMTS13 function, and 5 out of 6 engineered glycans on the Cys-rich domain also had no effect on ADAMTS13 function. However, a glycan attached at position 476 appreciably reduced both VWF binding and proteolysis. Substitution of Cys-rich sequences for the corresponding regions in ADAMTS1 identified a hydrophobic pocket involving residues Gly471-Val474 as being of critical importance for both VWF binding and proteolysis. Substitution of hydrophobic VWF A2 domain residues to serine in a region (residues 1642-1659) previously postulated to interact with the Cys-rich domain revealed the functional importance of VWF residues Ile1642, Trp1644, Ile1649, Leu1650, and Ile1651. Furthermore, the functional deficit of the ADAMTS13 Cys-rich Gly471-Val474 variant was dependent on these same hydrophobic VWF residues, suggesting that these regions form complementary binding sites that directly interact to enhance the efficiency of the proteolytic reaction.
Introduction
ADAMTS13 is a plasma metalloprotease that proteolytically regulates von Willebrand factor (VWF), a critical mediator of platelet tethering and primary hemostasis.1 VWF circulates in blood in multimeric forms of highly variable size, ranging from dimers to species that may exceed 60-mers.2 The multimeric size of VWF is central to its platelet-tethering function, with larger multimers conferring greater hemostatic potential than smaller forms.3,4 VWF function is regulated in plasma by ADAMTS13-mediated proteolysis, which under conditions of elevated shear forces converts VWF into smaller, less hemostatically active fragments.1 The physiological importance of the ADAMTS13-VWF axis is highlighted by cases of either inherited or acquired ADAMTS13 deficiency, which is associated with life-threatening thrombotic thrombocytopenic purpura.5 Moreover, high VWF and low ADAMTS13 plasma levels are independent risk factors for myocardial infarction and stroke.6,7
In free circulation, VWF multimers generally adopt a globular fold that not only shields the platelet-binding site in its A1 domain but also conceals both the scissile bond and high-affinity binding sites for ADAMTS13 within the core of the VWF A2 domain.8,9 This makes globular VWF ostensibly unable to bind platelets and also resistant to ADAMTS13-mediated proteolysis. However, during secretion of VWF from endothelial cells, as well as in other scenarios in which VWF is exposed to high shear stress (including at sites of vessel damage), VWF multimers unravel to expose the A1 domain that can potentiate platelet binding. Platelet capture can be concomitantly controlled through the unfolding of the central A2 domain.10,11 This process facilitates the exposure of cryptic binding sites in the VWF A2 domain that enable multiple specific interactions with ADAMTS13.1 These interactions function like a molecular zipper that presents the Tyr1605-Met1606 scissile bond in VWF to the ADAMTS13 active site.1 Of the reported binding interactions between the 2 molecules, the one between the ADAMTS13 spacer domain (involving Arg568, Arg660, Tyr661, and Tyr665) and the C terminus of the VWF A2 domain (involving Glu1660-Arg1668) is thought to be of highest affinity.12-17 This interaction approximates the 2 molecules, enabling an exosite in the ADAMTS13 disintegrin-like (Dis) domain (involving Arg349, Leu350, and Val352) to interact with residues near the VWF scissile bond (including Asp1614).17,18 This positions the scissile bond near the catalytic site, in turn allowing critical low-affinity interactions between the ADAMTS13 metalloprotease (MP) domain and the P3 residue of VWF (Leu1603) that precede accommodation of the P1-P1′ residues in VWF (Tyr1605-Met1606) into the S1 and S1′ pockets.19-21 The importance of the multiple exosites of ADAMTS13 in VWF proteolysis is exemplified by the finding that the MP domain alone cannot cleave VWF specifically. However, the addition of the domains C terminal to the MP domain progressively increases the efficiency of substrate proteolysis.18,22,23 Whereas functional contributions of the MP, Dis, and spacer domains to VWF proteolysis have been characterized, questions remain regarding the precise role that the cysteine-rich (Cys-rich) domain fulfils. Based on the crystal structure of the ADAMTS13 Dis to spacer fragment (termed DTCS) and models of the MP domain, in conjunction with knowledge of the location of VWF binding sites in the MP, Dis, and spacer domains (supplemental Figure 1 available at the Blood Web site), the Cys-rich domain, particularly the surface of the domain that is located between the Dis and spacer domain exosites, is well situated to interact directly with VWF and contribute to substrate proteolysis.1,17 In support of this contention are the findings by Gao et al, who compared the activities of C-terminally truncated ADAMTS13 mutants with and without the Cys-rich domain (termed MDTC and MDT, respectively). They found a 10-fold reduction in kcat/Km following removal of the Cys-rich domain,23 suggesting that the Cys-rich domain is, in addition to the spacer and the Dis domains, of critical importance for VWF regulation. Interestingly, when VWF substrates were used that lack the region Ile1642-Arg1659, MDTC and MDT proteolyzed these at a similar rate, which suggests that the function of the Cys-rich domain is dependent on VWF residues located within this region.23 We therefore examined how the Cys-rich domain functions at a molecular level.
Methods
Expression of recombinant ADAMTS13 Cys-rich domain variants
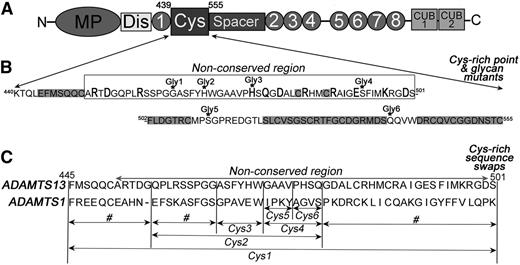
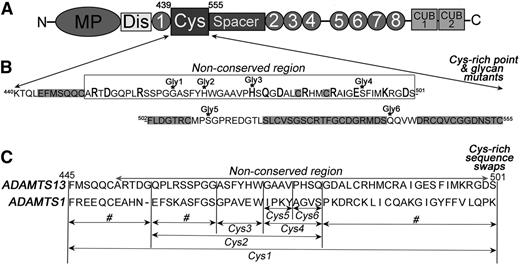
The wild-type (WT) human ADAMTS13 mammalian expression vector that fuses a myc-His tag to the C terminus has been described previously.24 Single point and composite mutants (R452A, D454A, R459A, H476A, H476A/Q478A, D480A, R484A, I490A, E492A, K497A, and D500A) and introduction of the N-linked glycan attachment motif mutants G464N (Gly1), H469N/G471T (Gly2), H476N/Q478T (Gly3), S493N/I495T (Gly4), S511N/P513T (Gly5), and Q539N/V541T (Gly6) were performed using KOD polymerase (Merck). ADAMTS13 Cys-rich domain swap variants in which specific sequences were replaced by the corresponding sequences from ADAMTS1 were also generated by polymerase chain reaction, including Phe445-Ser501 (Cys1), Phe445-Gly455, Gln456-Gly464, Gln456-Gln478 (Cys2), Gly479-Ser501, Ala465-Trp470 (Cys3), Gly471-Gln478 (Cys4), Gly471-Val474 (Cys5), and Pro475-Gln478 (Cys6) (Figure 1).
Cys-rich domain amino acid sequence and sites of mutagenesis. (A) Domain organization of ADAMTS13. The Cys-rich domain comprises amino acids Lys440-Cys555. (B) The amino acids sequence of the Cys-rich domain contains a region that is highly conserved among ADAMTS family members (highlighted in gray) and a nonconserved region (indicated). In the nonconserved region, charged amino acids were individually mutated to alanine (bold and larger font). Amino acids that were mutated to asparagine for the introduction of an NXT glycosylation motif are indicated (Gly1-6). See supplemental Figure 2 for the location of each variant on a model of ADAMTS13. (C) Cys-rich domain sequence swap variants were generated by substitution of selected sequences for the corresponding sequence in ADAMTS1. Variants named Cys1-6 were used for functional analysis. Those swap variants labeled # were generated and expressed, but not secreted.
Cys-rich domain amino acid sequence and sites of mutagenesis. (A) Domain organization of ADAMTS13. The Cys-rich domain comprises amino acids Lys440-Cys555. (B) The amino acids sequence of the Cys-rich domain contains a region that is highly conserved among ADAMTS family members (highlighted in gray) and a nonconserved region (indicated). In the nonconserved region, charged amino acids were individually mutated to alanine (bold and larger font). Amino acids that were mutated to asparagine for the introduction of an NXT glycosylation motif are indicated (Gly1-6). See supplemental Figure 2 for the location of each variant on a model of ADAMTS13. (C) Cys-rich domain sequence swap variants were generated by substitution of selected sequences for the corresponding sequence in ADAMTS1. Variants named Cys1-6 were used for functional analysis. Those swap variants labeled # were generated and expressed, but not secreted.
ADAMTS13 and ADAMTS13 variants were transiently expressed in HEK293T in OptiMEM (Invitrogen). After 3 days, conditioned medium was harvested and concentrated 10-fold using 50-kDa molecular weight cutoff centrifugal filter units (Millipore). Expression and secretion were analyzed by western blotting using an anti-myc antibody (Ab) (Santa Cruz) or biotinylated polyclonal anti-ADAMTS13 TSP(2-4) for detection. To assess successful addition of a novel glycan for the Gly1-6 mutants, conditioned medium was first treated with 350 nM thrombin (Enzyme Research Laboratories) overnight, and the cleavage fragment Ser394-Arg910 was detected by western blot with biotinylated polyclonal anti-ADAMTS13 TSP(2-4). The concentrations of ADAMTS13 and its variants were measured by enzyme-linked immunosorbent assay, as previously described.25
Expression and purification of VWF115, VWF115 mutants, and VWF106
Recombinant VWF A2 domain fragments, VWF115 (VWF residues 1554-1668),26 and its C-terminally truncated variant VWF106 (residues 1554-1659)14 were used as substrates for ADAMTS13 and also to measure ADAMTS13 binding. VWF115 and VWF106 both contain an N-terminal 6xHis and Xpress epitope tag. They were expressed in Rosetta DE-3 E. coli (Novagen), purified, and quantified as described previously.26 The mutations I1649S/L1650S/I1651S, termed 3S, and I1642S/W1644S/I1649S/L1650S/I1651S, termed 5S, were introduced by site-directed mutagenesis using KOD Hot-Start polymerase (Merck) and confirmed by sequencing.
ADAMTS13 activity assays
ADAMTS13 activity was analyzed using VWF115 or VWF115 3S and 5S variants as a substrate as previously described.18 As indicated, 1 to 45 nM ADAMTS13 or ADAMTS13 variant in concentrated conditioned media was preincubated with 5 mM CaCl2 for 1 hour before the addition of 6 µM VWF115 or VWF115 variant (both final concentrations) to start the reaction. Subsamples were removed from 0- to 2-hour time points, stopped with EDTA, and proteolysis analyzed qualitatively by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining. All assays were repeated at least 3 times, and representative gels are shown. To derive kcat/Km for VWF115(5S), activity assays were carried out as above, except that 1 µM substrate was used and proteolysis was quantified by densitometry (ImageJ) of western blots using an anti-Xpress antibody (Life Tech). Time-course reaction curves (n = 3-6) were plotted using GraphPad Prism and kcat/Km derived as described previously.26
When the fluorogenic substrate, FRETS-VWF73, was used, 100 µL WT ADAMTS13 or ADAMTS13 variants in conditioned medium diluted to 0.6 nM in assay buffer (20 mM Tris, 25 mM CaCl2, and 0.005% Tween-20 at pH 7.5) was added to a white 96-well plate. A total of 100 µL FRETS-VWF73 substrate was added to each well to start the reaction (final concentration of 1 µM). Fluorescence (excitation 340 nm; emission 460 nm) was measured at 37°C at 90-second intervals for 1 hour using a Fluostar Omega plate reader (BMG Labtech). To derive kcat/Km, assays were repeated as above, except that ADAMTS13 and ADAMTS13 variant concentrations were adjusted for each mutant to enable complete proteolysis to occur within 90 minutes.
Plate-binding assays
Binding of ADAMTS13 and ADAMTS13 variants to immobilized recombinant VWF115 was analyzed as described previously.27 Briefly, purified VWF115 or VWF115 3S, 5S variants were diluted to 50 or 100 nM (as indicated) in 50 mM sodium carbonate buffer (pH 9.6) and immobilized onto a 96-well plate overnight at 4°C. After washing and blocking the wells with phosphate-buffered saline (PBS)/0.1% Tween and PBS/3% bovine serum albumin, respectively, 100 µL of 0 to 200 nM WT ADAMTS13 or ADAMTS13 variant diluted in 1% bovine serum albumin/30 mM EDTA/PBS was added to wells and incubated for 2 hours. Thereafter, wells were washed and biotinylated anti-ADAMTS13 TSP(2-4) antibody and streptavidin/horseradish peroxidase used for detection. Equal coating of VWF115 and VWF115 mutants was confirmed using a mouse monoclonal Ab that detects the Xpress tag that is fused to VWF115.
Results
Sequence analysis of the ADAMTS13 Cys-rich domain
Based upon amino acid sequence alignments, the Cys-rich domain contains a conserved region (spanning Phe502 to Cys555) that is broadly conserved among ADAMTS family members (supplemental Figure 2). This conservation points to a structural role for this region in the global fold of the domain. Consistent with this contention, this conserved region sits between the adjacent TSP1 and spacer domains in the crystal structure of these domains and is not exposed to the substrate-binding face of the protease. There is also a nonconserved region in the Cys-rich domain (spanning Ala451 to Ser501) with much higher sequence variability among ADAMTS family members, suggesting that this region may be involved in enzyme-specific functions. As this nonconserved Cys-rich region is located on the VWF-binding face of ADAMTS13, we hypothesized that it may contribute specifically to ADAMTS13 function.
Targeted mutagenesis of the ADAMTS13 Cys-rich domain
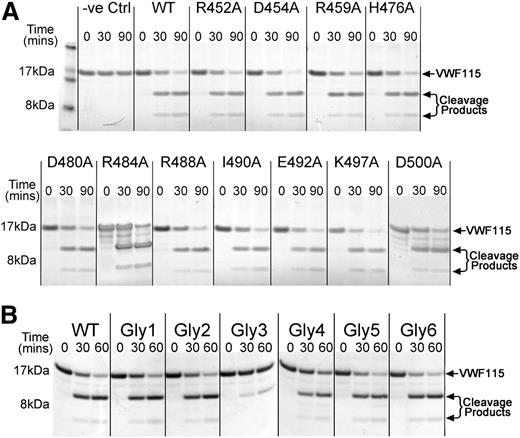
We first targeted the nonconserved region in the ADAMTS13 Cys-rich domain by site-directed mutagenesis (Figure 1A-B and supplemental Figure 2). Eleven predominantly charged and surface-exposed amino acids in this nonconserved region were mutated to alanine (Figure 1B and supplemental Figure 2). All mutants were expressed and secreted normally. To assess the influence of these substitutions on ADAMTS13 function, we assayed their activity using VWF115 as a substrate. These results revealed that all ADAMTS13 mutants cleaved VWF115 with similar efficiency to WT ADAMTS13 (Figure 2A), suggesting that none of the mutated residues have a major influence on VWF proteolysis or form part of any important functional exosite in ADAMTS13.
Functional analysis of ADAMTS13 Cys-rich domain point mutants and engineered glycan mutants. (A) Activity assays were set up in which 0.5 nM ADAMTS13 or ADAMTS13 point mutant in concentrated conditioned media was incubated with 6 µM VWF115. Control reactions containing media without ADAMTS13 were set up in parallel. At time points from 0 to 90 minutes, aliquots were removed, stopped with EDTA, and analyzed by SDS-PAGE and Coomassie staining. Uncleaved VWF115 is a 16.9-kDa protein. Cleavage products of 10 kDa and 6.9 kDa arising from specific ADAMTS13-mediated proteolysis are marked by arrows. (B) ADAMTS13 variants containing novel engineered glycans at different positions on the surface of the Cys-rich domain were generated and their ability to proteolyze VWF115 assessed. The analysis was done as in panel A, except that 1 nM ADAMTS13 or variant was used and the third time point was taken at 60 minutes.
Functional analysis of ADAMTS13 Cys-rich domain point mutants and engineered glycan mutants. (A) Activity assays were set up in which 0.5 nM ADAMTS13 or ADAMTS13 point mutant in concentrated conditioned media was incubated with 6 µM VWF115. Control reactions containing media without ADAMTS13 were set up in parallel. At time points from 0 to 90 minutes, aliquots were removed, stopped with EDTA, and analyzed by SDS-PAGE and Coomassie staining. Uncleaved VWF115 is a 16.9-kDa protein. Cleavage products of 10 kDa and 6.9 kDa arising from specific ADAMTS13-mediated proteolysis are marked by arrows. (B) ADAMTS13 variants containing novel engineered glycans at different positions on the surface of the Cys-rich domain were generated and their ability to proteolyze VWF115 assessed. The analysis was done as in panel A, except that 1 nM ADAMTS13 or variant was used and the third time point was taken at 60 minutes.
Engineering novel N-linked glycans into ADAMTS13 identifies a functional region within the Cys-rich domain
As a further search for functionally important sites, we screened larger areas of the Cys-rich domain through the introduction of N-linked glycan attachment motifs (N-X-S/T) by site-directed mutagenesis (Figure 1B and supplemental Figure 2). As N-linked glycans are ∼5 kDa and have a diameter of ∼30Å they have the potential to disrupt interactions in the vicinity of the attachment site. We designed 8 mutants carrying a new NXS/T motif at various sites in the Cys-rich domain predicted to be located on the surface of the domain. All were expressed and secreted efficiently. Of the 8 variants, we confirmed addition of a new glycan in 6 of these, as visualized by a band shift in a western blot of conditioned media (supplemental Figure 3). We assessed the ability of these 6 ADAMTS13 glycan mutants (termed Gly1-6) to cleave VWF115. All variants proteolyzed VWF115 efficiently, except Gly3 (H476N/Q478T), for which greatly reduced proteolysis was observed (Figure 2B). This suggests that the new glycan at position 476 disrupts a functionally important interaction between ADAMTS13 and VWF or obstructs the path of VWF between interaction sites on ADAMTS13. This effect was very likely attributable to the insertion of a new glycan, as when H476 and Q478 were substituted to alanine, there was no effect upon VWF115 proteolysis (not shown).
ADAMTS13 Cys-rich domain swap variants identify Gly471-Val474 as important to ADAMTS13 function
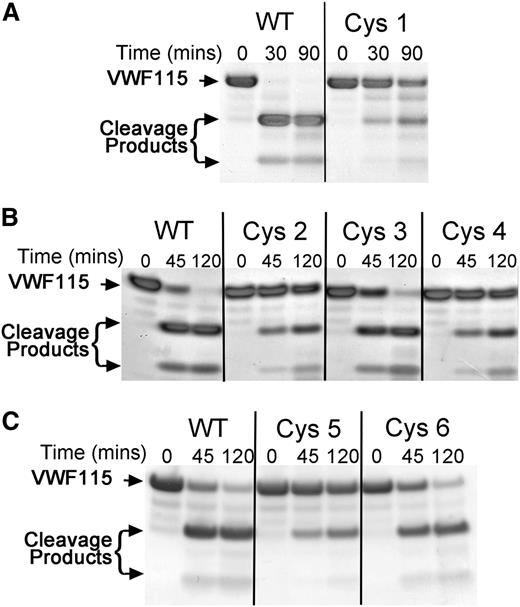
The next approach that we used to identify functionally important sites involved swapping selected sequences in the nonconserved region in the ADAMTS13 Cys-rich domain with the corresponding sequence in ADAMTS1 (Figure 1C). Of the 9 swap variants that were made, 6 (termed Cys1-6) were secreted efficiently and therefore analyzed functionally. In the ADAMTS13 Cys1 variant, Phe445-Ser501 was replaced, which spans the entire nonconserved region. Using 5 nM WT ADAMTS13, VWF115 proteolysis went to completion in <30 minutes. However, under these same conditions, the proteolysis of VWF115 by the Cys1 variant was appreciably reduced (Figure 3A), and even after 90 minutes, VWF115 was still only partially proteolyzed. This suggests that the nonconserved region within the ADAMTS13 Cys-rich domain is of functional importance for ADAMTS13.
Functional analysis of ADAMTS13 Cys-rich domain sequence swap variants. (A) A total of 5 nM ADAMTS13 or ADAMTS13 Cys1 variant in conditioned medium was incubated with 6 µM VWF115. Aliquots were taken and stopped with EDTA at 0, 30, and 90 minutes and analyzed by SDS-PAGE and Coomassie staining. Uncleaved VWF115 and cleavage products arising from specific ADAMTS13-mediated proteolysis are marked by arrows. (B) ADAMTS13 variants Cys2 to Cys4 were analyzed as in panel A, except that 2 nM ADAMTS13 was used and aliquots were taken at 0, 45, and 120 minutes. (C) ADAMTS13 variants Cys5 and Cys6 were analyzed as in panel B.
Functional analysis of ADAMTS13 Cys-rich domain sequence swap variants. (A) A total of 5 nM ADAMTS13 or ADAMTS13 Cys1 variant in conditioned medium was incubated with 6 µM VWF115. Aliquots were taken and stopped with EDTA at 0, 30, and 90 minutes and analyzed by SDS-PAGE and Coomassie staining. Uncleaved VWF115 and cleavage products arising from specific ADAMTS13-mediated proteolysis are marked by arrows. (B) ADAMTS13 variants Cys2 to Cys4 were analyzed as in panel A, except that 2 nM ADAMTS13 was used and aliquots were taken at 0, 45, and 120 minutes. (C) ADAMTS13 variants Cys5 and Cys6 were analyzed as in panel B.
The ADAMTS13 Cys2 variant, in which Gln456-Gln478 was replaced, was analyzed under slightly different conditions. Using 2 nM WT ADAMTS13, proteolysis of VWF115 was almost complete after 45 minutes, whereas the Cys2 variant was still only partially proteolyzed after 120 minutes (Figure 3B). Interestingly, this region of 23 amino acids contains the site where the glycan is attached in the Gly3 variant.
The ADAMTS13 variant Cys3 (Ala465-Trp470) proteolyzed VWF115 near normally, whereas variant Cys4 (Gly471-Gln478) had a functional deficit similar to Cys2 (Figure 3B). Finally, we generated 2 further ADAMTS13 variants, Cys5 and Cys6, in which Gly471-Val474 and Pro475-Gln478, respectively, were replaced. Proteolysis of VWF115 by variant Cys6 was minimally affected (Figure 3C), suggesting that the residues Pro475-Gln478 are not of major importance. Conversely the Cys5 variant exhibited a reduced rate of VWF115 proteolysis similar to Cys1, Cys2, and Cys4, suggesting that it is substitution of this 4-amino-acid region that is responsible for the reduced activity (Figure 3C).
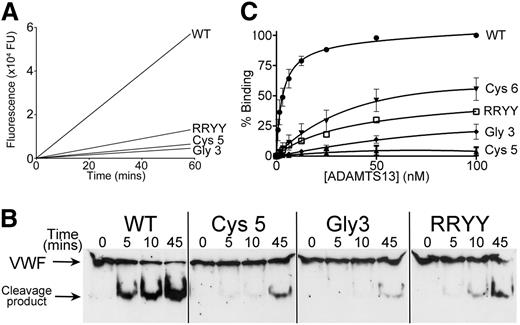
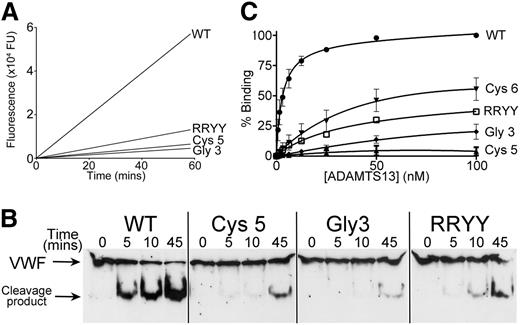
To confirm and quantify the reductions in activity of the Cys5 and the Gly3 variants, we used the fluorogenic ADAMTS13 substrate FRETS-VWF73 (Figure 4A and Table 1). This revealed that the kcat/Km for the proteolysis of FRETS-VWF73 by WT ADAMTS13 (9.1 ± 1.5 × 105 M−1s−1) was very similar to previous reports.13,15,23 By comparison, the kcat/Km for ADAMTS13 Cys5 was reduced 6-fold (1.5 ± 0.4 × 105 M−1s−1), whereas the ADAMTS13 Gly3 mutant exhibited an 11-fold (0.8 ± 0.1 × 105 M−1s−1) reduction in catalytic efficiency. For comparison, we included in this assay a spacer domain variant, termed RRYY, containing the R568A/R660A/Y661A/Y665A mutations that have previously been reported to disrupt a region of the spacer domain that is critical to VWF binding.14,17 In this assay, by comparison with WT ADAMTS13, ADAMTS13 RRYY had a kcat/Km for VWF73 proteolysis that was 5-fold reduced (1.8 ± 0.2 × 105 M−1s−1). When compared with the 6-fold reduction of the Cys5 variant, this provides an indication of the relative importance of the exosites that are disrupted in the Cys5 and Gly3 variants.
Analysis of ADAMTS13 Cys5 and ADAMTS13 Gly3 variants function. (A) FRETS-VWF73 was used to quantify the activity of ADAMTS13 variants Gly3 and Cys5 and compare it to the residual activity of spacer domain mutant RRYY. A total of 0.6 nM WT ADAMTS13 or ADAMTS13 variant in conditioned medium was incubated with 2 µM FRETS-VWF73 and fluorescence measured over time. A representative graph is shown (n = 5). (B) Proteolysis of recombinant full-length VWF (N1602A/C1669G/C1670G) by 8 nM ADAMTS13 or ADAMTS13 variants. Subsamples were stopped with EDTA at designated time points and analyzed by western blotting under reducing conditions using an anti-VWF monoclonal Ab that detects both full-length VWF and the 176-kDa cleavage product. (C) Binding of WT ADAMTS13 and ADAMTS13 Cys-rich Cys5, Cys6, and Gly3 variants to VWF115. VWF115 (50 nM) was immobilized on microtiter wells and incubated with increasing concentrations of ADAMTS13 or variant. Wells were washed and ADAMTS13 binding detected using a biotinylated anti-ADAMTS13 polyclonal antibody followed by streptavidin/horseradish peroxidase and orthophenylenediamine. Absorbance at 492 nm was used to quantify binding. Mean values and error bars are shown (n = 3) except for variant RRYY (n = 1).
Analysis of ADAMTS13 Cys5 and ADAMTS13 Gly3 variants function. (A) FRETS-VWF73 was used to quantify the activity of ADAMTS13 variants Gly3 and Cys5 and compare it to the residual activity of spacer domain mutant RRYY. A total of 0.6 nM WT ADAMTS13 or ADAMTS13 variant in conditioned medium was incubated with 2 µM FRETS-VWF73 and fluorescence measured over time. A representative graph is shown (n = 5). (B) Proteolysis of recombinant full-length VWF (N1602A/C1669G/C1670G) by 8 nM ADAMTS13 or ADAMTS13 variants. Subsamples were stopped with EDTA at designated time points and analyzed by western blotting under reducing conditions using an anti-VWF monoclonal Ab that detects both full-length VWF and the 176-kDa cleavage product. (C) Binding of WT ADAMTS13 and ADAMTS13 Cys-rich Cys5, Cys6, and Gly3 variants to VWF115. VWF115 (50 nM) was immobilized on microtiter wells and incubated with increasing concentrations of ADAMTS13 or variant. Wells were washed and ADAMTS13 binding detected using a biotinylated anti-ADAMTS13 polyclonal antibody followed by streptavidin/horseradish peroxidase and orthophenylenediamine. Absorbance at 492 nm was used to quantify binding. Mean values and error bars are shown (n = 3) except for variant RRYY (n = 1).
As VWF73 and VWF115 are short VWF A2 domain fragments, their proteolysis by ADAMTS13 may not be truly reflective of the cleavage of full-length VWF multimers. We therefore compared the ability of WT ADAMTS13 and the ADAMTS13 Cys5 and Gly3 variants to cleave full-length VWF and visualized proteolysis by western blot under reducing conditions. Because the chaotropic reagents that are commonly used to unfold VWF in vitro under static conditions disrupt hydrophobic interactions28 and may interfere with Cys-rich domain binding to VWF, we used a recombinant VWF variant (N1602A/C1669G/C1670G) that is cleaved efficiently in the absence of denaturants due to mutation of both the A2 domain Ca2+-binding site and the 2 vicinal cysteines at the C-terminal end of the A2 domain.29 This full-length VWF variant was rapidly cleaved in the presence of 8 nM WT ADAMTS13 and approached completion after 45 minutes (Figure 4B), whereas only limited proteolysis was seen at this time for the ADAMTS13 variants Cys5, Gly3, and RRYY, confirming the reduced activity of these ADAMTS13 variants seen when short substrates VWF73 and VWF115 were used.
ADAMTS13 Cys-rich domain variants exhibit reduced VWF binding
To explore the mechanism underlying the reduced activity of the ADAMTS13 Gly3 and Cys5 variants, we analyzed the binding affinity of ADAMTS13 Gly3, Cys5, and Cys6 for VWF115 using a plate-binding assay. Using this approach, WT ADAMTS13 bound VWF115 with low nanomolar affinity (Figure 4C) similar to previous reports.26,30 The binding of the variants Gly3 and Cys5 to VWF115 was greatly reduced compared with WT ADAMTS13, whereas binding of variant Cys6 was moderately affected. Derivation of KD values for these interactions was not possible due to the large reduction in binding, which did not approach saturation. This assay monitors the tight binding interaction between ADAMTS13 and the unraveled VWF A2 domain and has previously been considered to be highly dependent upon the ADAMTS13 spacer domain. This contention was further corroborated by the marked reduction in VWF115 binding by the spacer domain variant RRYY.
Identification of hydrophobic VWF A2 domain residues that contribute to ADAMTS13 binding
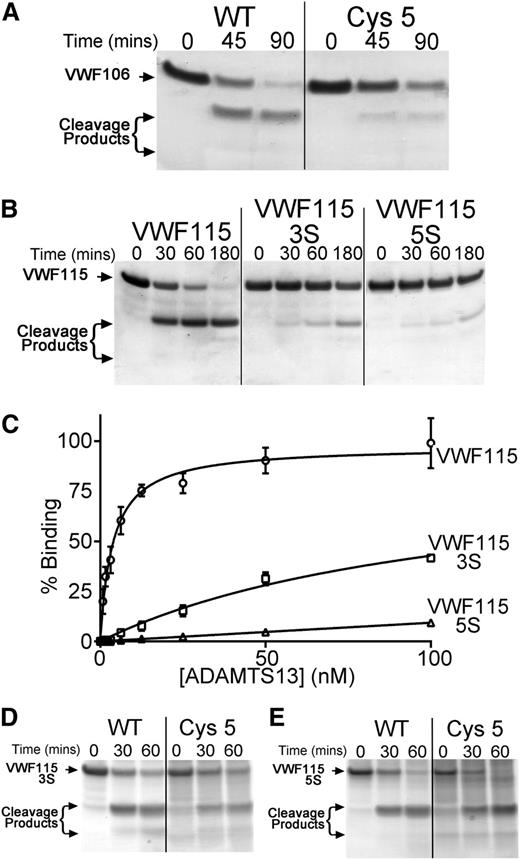
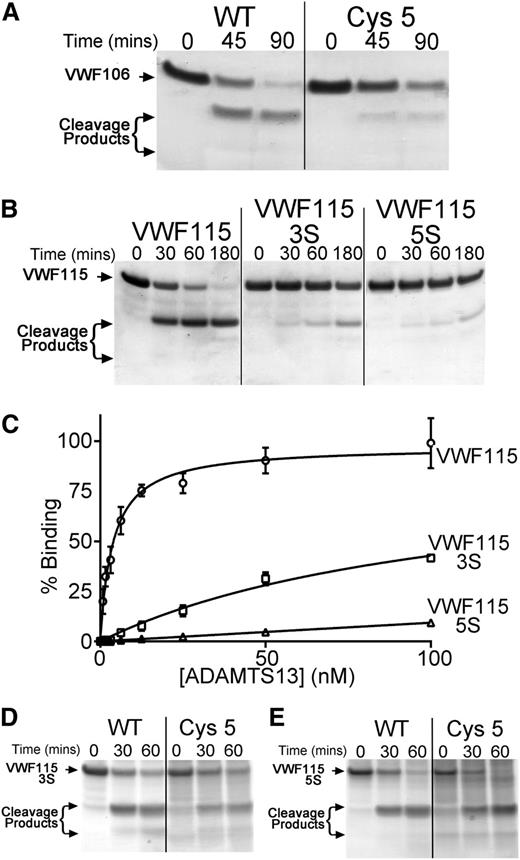
Inspection of the crystal structure of the ADAMTS13 Cys-rich domain (Protein Data Bank ID 3GHM)17 suggested that a hydrophobic pocket in the Cys-rich domain (involving Ala472, Ala473, and Val474) may have been disrupted in the Cys5 variant that might represent a distinct exosite to that in the spacer domain or, alternatively, may be a part of an extended exosite that includes the spacer domain. To examine this further, we analyzed the proteolysis of VWF106, a recombinant VWF A2 domain fragment that is identical to VWF115 but lacks the 9 C-terminal amino acids previously reported to be essential for spacer domain binding. Proteolysis of this substrate is appreciably less efficient than that of VWF115. Consequently, we used 17 nM ADAMTS13 in these reactions. Under these conditions, VWF106 was almost completely proteolyzed after 60 minutes (Figure 5A). The Cys5 variant still exhibited a proteolytic deficit, suggesting that the effect of the substitutions in Cys5 may be distinct from the function of the spacer domain (Figure 5A).
Identification of hydrophobic VWF A2 domain residues that contribute to ADAMTS13 binding. (A) A total of 6 µM VWF106 (Glu1554-Arg1659) was incubated with WT ADAMTS13 or ADAMTS13 Cys5 (17 nM) in conditioned medium, and aliquots taken at the indicated time points were analyzed by SDS-PAGE and Coomassie staining. (B) VWF115(3S) and VWF115(5S) containing composite mutations of hydrophobic residues to serine were used in functional analyses as in panel A, except that 1 nM WT ADAMTS13 was used. (C) Binding of WT ADAMTS13 to VWF115(3S) and (5S). VWF115 or variant (100 nM) was immobilized on microtiter wells, and binding was assessed as in Figure 4B. Mean values ± SD are shown (n = 3). (D-E) Proteolysis of VWF115(3S) (D) and VWf115(5S) (E) by 24 nM WT ADAMTS13 and ADAMTS13 Cys5. Cleavage was analyzed as in panel A-B.
Identification of hydrophobic VWF A2 domain residues that contribute to ADAMTS13 binding. (A) A total of 6 µM VWF106 (Glu1554-Arg1659) was incubated with WT ADAMTS13 or ADAMTS13 Cys5 (17 nM) in conditioned medium, and aliquots taken at the indicated time points were analyzed by SDS-PAGE and Coomassie staining. (B) VWF115(3S) and VWF115(5S) containing composite mutations of hydrophobic residues to serine were used in functional analyses as in panel A, except that 1 nM WT ADAMTS13 was used. (C) Binding of WT ADAMTS13 to VWF115(3S) and (5S). VWF115 or variant (100 nM) was immobilized on microtiter wells, and binding was assessed as in Figure 4B. Mean values ± SD are shown (n = 3). (D-E) Proteolysis of VWF115(3S) (D) and VWf115(5S) (E) by 24 nM WT ADAMTS13 and ADAMTS13 Cys5. Cleavage was analyzed as in panel A-B.
Gao et al previously reported that VWF A2 domain residues Gln1624-Arg1641 can be deleted from short A2 domain fragment substrates without a major reduction in the rate of proteolysis by ADAMTS13.23 This prompted us to search VWF residues Ile1642-Arg1659 for a complementary hydrophobic exosite that might interact with the ADAMTS13 Cys-rich domain. Within this VWF region, there are several hydrophobic residues that could potentially mediate a hydrophobic interaction with ADAMTS13 (supplemental Figure 1). To investigate this, we generated and expressed the composite mutants VWF115(3S) (I1649S/L1650S/I1651S) and VWF115(5S) (I1642S/W1644S/I1649S/L1650S/I1651S) in which clusters of these hydrophobic residues were substituted for serine. We purified and quantified these composite mutants and analyzed their proteolysis by ADAMTS13. Interestingly, using 1 nM WT ADAMTS13, VWF115 was proteolyzed to completion after 60 minutes, whereas VWF115(3S) was cleaved appreciably slower and VWF115(5S) was cleaved even less efficiently, with the majority of each substrate remaining uncleaved even after 3 hours (Figure 5B). To quantify proteolysis of VWF115 and VWF115(5S), ADAMTS13 concentration was titrated for each substrate to identify a concentration at which proteolysis reached completion within 90 minutes (2.5 nM for VWF115 and 24 nM for VWF115[5S]). The kcat/Km was derived from time-course curves (supplemental Figure 4) based on densitometric quantitation of western blots of the cleavage reactions using an anti-Xpress antibody. This revealed that the kcat/Km of VWF115 proteolysis by WT ADAMTS13 was (3.2 ± 1.5 × 105 M−1s−1, n = 4), which was very similar to previous reports.18 However, the kcat/Km for VWF115(5S) proteolysis was reduced 12-fold (0.26 ± 0.07 × 105 M−1s−1, n = 3).
The reduced proteolysis of the VWF115(3S) and (5S) mutants is accompanied by a marked effect on ADAMTS13 binding, as determined using the plate-binding assay (Figure 5C). WT ADAMTS13 bound to VWF115 with high affinity, as before. The binding of ADAMTS13 to VWF115(3S) was markedly reduced and to VWF(5S) was even further reduced. Again, derivation of KD values for the interaction of WT ADAMTS13 with VWF115(3S) and (5S) was not possible. These results suggest that the 5 hydrophobic VWF residues Ile1642, Trp1644, Ile1649, Leu1650, and Ile1651 are involved in hydrophobic interactions between ADAMTS13 and VWF.
As these hydrophobic VWF residues are in a region that has previously been proposed to interact with the ADAMTS13 Cys-rich domain, and because of the finding that residues Gly471-Val474 within the Cys-rich domain may form a hydrophobic pocket that is critical for ADAMTS13 activity, we hypothesized that these ADAMTS13 and VWF residues are involved in complementary hydrophobic interactions. To examine this, we compared the proteolysis of VWF115(3S) and VWF115(5S) by WT ADAMTS13 and ADAMTS13 Cys5. Interestingly, VWF115(3S) and VWF115(5S) were proteolyzed at very similar rates by both WT and Cys5 (Figure 5D-E and supplemental Figure 4). The kcat/Km for VWF115(5S) proteolysis by ADAMTS13 Cys5 was estimated to be 0.24 ± 0.08 × 105 M−1s−1 (n = 3), which is very similar to that derived for WT ADAMTS13 (0.26 ± 0.07 × 105 M−1s−1, n = 3). As the functional difference between WT ADAMTS13 and ADAMTS13 Cys5 was lost using VWF115(3S) or VWF115(5S), this implies that the deficit in Cys5 is dependent upon VWF residues Ile1642, Trp1644, Ile1649, Leu1650, and Ile1651.
Discussion
The MP domain of ADAMTS13 requires VWF binding exosites in the other domains to enable its proteolysis of VWF. The 2 exosites that have been reported by multiple research groups involve (1) the ADAMTS13 Dis domain residues Arg349 and Leu35017,18 and (2) the ADAMTS13 spacer domain residues Arg568, Arg660, Tyr661, and Tyr665.14,15,17 The distance between these 2 exosites is >100 Å in the crystal structure of ADAMTS13 DTCS, which has raised the question of whether there are additional exosites in the intervening TSP-1 or Cys-rich domains. Although previous reports have suggested that the Cys-rich domain directly interacts with VWF,17,22,23 this is the first comprehensive analysis of this domain that identifies its functional importance.
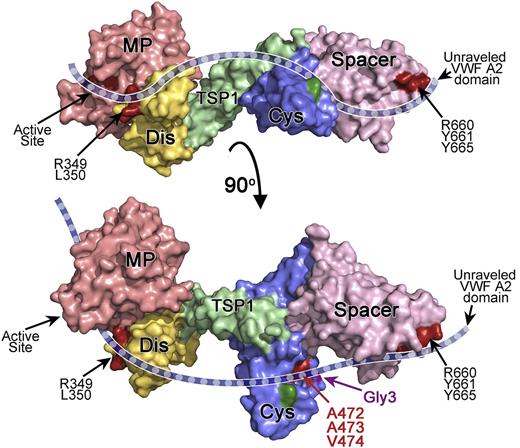
The results of our analysis support an important contribution of the ADAMTS13 Cys-rich domain to VWF binding and proteolysis. We show that out of 6 engineered glycans at various positions on the surface of the Cys-rich domain, only 1 affected VWF binding and proteolysis. The position of this glycan (residue 476) gives an indication of how the unraveled VWF A2 domain bridges the 2 known exosites in the spacer and Dis domains. Our initial alanine scanning of the Cys-rich domain did not identify any amino acids that are critical for efficient proteolysis of VWF. However, this approach was limited to charged residues and, therefore, hydrophobic residues were not investigated. Our subsequent screening using sequence swap variants suggested a functional role for amino acids located in a hydrophobic pocket comprising amino acids Gly471-Val474 (Figure 6). This pocket is situated on the same face as the spacer domain exosite (Figure 6) and in the vicinity of the attachment site of the engineered glycan (Gly3) that also dramatically reduced VWF binding (Figure 4A).
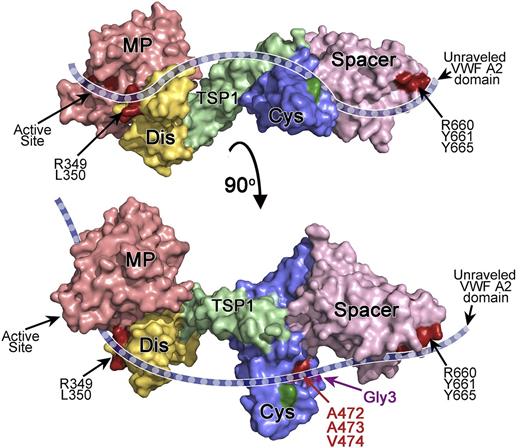
Proposed model for the interaction of ADAMTS13 with the unraveled VWF A2 domain. Depicted is a model of ADAMTS13 MDTCS. The model is based on the crystal structure of ADAMTS13 DTCS17 and a homology model of MP-Dis. Surface representation is depicted and color-coded as follows: MP (red), Dis (yellow), TSP1 (green), Cys-rich (blue), and spacer (pink). The bottom figure has been rotated ∼90° compared with the top figure. The active site, Dis exosite (R349 and L350), and spacer exosite (R660, Y661, and Y665) are shown in dark red and labeled. A cartoon representation of unraveled VWF A2 domain (dashed blue ribbon) is shown to indicate how we hypothesize it extends across the ADAMTS13 active site and the exosites in the ancillary Dis, Cys-rich, and spacer domains, based on the results of mutagenesis studies. The location of the hydrophobic pocket in the Cys-rich domain involving A472, A473, and V474 is shown in red. The position of the novel glycan attachment site that impairs VWF proteolysis (Gly3) is shown in purple. In dark green is the location of the Gly2 variant glycan attachment site that had no effect on proteolysis.
Proposed model for the interaction of ADAMTS13 with the unraveled VWF A2 domain. Depicted is a model of ADAMTS13 MDTCS. The model is based on the crystal structure of ADAMTS13 DTCS17 and a homology model of MP-Dis. Surface representation is depicted and color-coded as follows: MP (red), Dis (yellow), TSP1 (green), Cys-rich (blue), and spacer (pink). The bottom figure has been rotated ∼90° compared with the top figure. The active site, Dis exosite (R349 and L350), and spacer exosite (R660, Y661, and Y665) are shown in dark red and labeled. A cartoon representation of unraveled VWF A2 domain (dashed blue ribbon) is shown to indicate how we hypothesize it extends across the ADAMTS13 active site and the exosites in the ancillary Dis, Cys-rich, and spacer domains, based on the results of mutagenesis studies. The location of the hydrophobic pocket in the Cys-rich domain involving A472, A473, and V474 is shown in red. The position of the novel glycan attachment site that impairs VWF proteolysis (Gly3) is shown in purple. In dark green is the location of the Gly2 variant glycan attachment site that had no effect on proteolysis.
Physiologically, the proteolysis of VWF by ADAMTS13 requires VWF to unfold in response to elevated shear stress in the circulation, as the binding sites for ADAMTS13, as well as the scissile bond itself, are otherwise not solvent exposed. A consequence of VWF unfolding is the exposure of hydrophobic residues that are buried inside the domain when VWF circulates in plasma in its globular form. Our results suggest that some of these hydrophobic residues (ie, Ile1642, Trp1644, Ile1649, Leu1650, and Ile1651) contribute to ADAMTS13 binding, as mutagenesis to serine in VWF115(3S) and VFW115(5S) resulted in appreciably reduced binding and proteolysis. These hydrophobic VWF residues may directly interact with the hydrophobic pocket in the ADAMTS13 Cys-rich domain. This contention was supported by the finding that whereas the Cys5 mutant cleaved VWF115 poorly compared with WT ADAMTS13, it cleaved the VFW115(5S) mutant at a similar rate as WT ADAMTS13 (Figure 5D), suggesting that the functional deficit is dependent on these hydrophobic VWF residues.
By truncating VWF substrates as well as making internal deletions, Gao et al previously showed that VWF residues located within Ile1642-Arg1659 and Glu1660-Arg1668 are necessary for efficient proteolysis, the contribution of Glu1660-Arg1668 being dependent on the spacer domain.23 Wu et al demonstrated that individual mutagenesis of the 6 charged residues within the region Ile1642-Arg1668 resulted in a 2-fold reduction in proteolysis for 2 of the 6 residues, whereas there was no effect for the other 4.16 These 2 charged residues therefore fail to account for the reported 75- to 200-fold reduction in proteolysis when the whole region was deleted. Based on our results, we propose that hydrophobic binding interactions may represent an important component of this region’s interaction with ADAMTS13.
Gao et al have also shown that when the region Gln1624-Arg1641 is deleted from VWF73, it does not appreciably alter the rate of proteolysis.23 In this deletion mutant, the maximal distance between Asp1614, which interacts with Arg349 in the Dis domain of ADAMTS13, and Ile1649 is ∼87Å, ie, when this VWF fragment is fully extended. From this we can conclude that any ADAMTS13 residues that may bind to Ile1649, Leu1650, and Ile1651 should be located within 87 Å of Arg349, which most Cys-rich domain residues are in the crystal structure of ADAMTS13 DTCS, but spacer domain exosite residues are not.
The functionally important Cys-rich residues that we identified (Gly471-Val474) lie in proximity to residues that have previously been reported to be important for efficient proteolysis of VWF, including Pro475.17,31 The East Asian–specific polymorphism P475S causes an ∼16% reduction in plasma activity, which is consistent with the functional importance of the adjacent Gly471-Val474.
Another interesting aspect of our results is that for our ADAMTS13 variant in which Gly471-Val474 was replaced (Cys5), very little residual binding remained in our assay, which could mean that binding of the spacer domain to VWF may be dependent on the function of the Cys-rich domain.
Jian et al demonstrated that an ADAMTS13 mutant with spacer domain mutations R660K/F592Y/R568K/Y661F/Y665F has increased activity.32 These mutations have recently been shown to disrupt an autoinhibitory binding between these VWF-binding residues in the spacer domain and the C-terminal domains of ADAMTS13.33 In WT ADAMTS13, this autoinhibition appears to be disrupted by conformational changes that occur upon binding to globular VWF. Whether the C-terminal domains also interfere with binding of the Cys-rich domain to VWF is unknown.33,34
In summary, our results demonstrate an important role for the Cys-rich domain in VWF proteolysis and identify hydrophobic residues in both ADAMTS13 and VWF that are critical for efficient proteolysis. These findings are in agreement with previous findings that suggested the presence of extensive contacts between exosites in ADAMTS13 and VWF, involving multiple domains. Although the mutagenesis studies that have identified exosites in ADAMTS13 have greatly advanced our understanding of VWF proteolysis, they need to be complemented with cocrystal structures of ADAMTS13 bound to VWF fragments to confirm that the reported amino acids do indeed form direct contacts with complementary exosites in VWF when the 2 molecules are bound.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge technical assistance from Matthew Jaring and Jordan Green and thank Chris Lynch (Imperial College London) for providing recombinant VWF (N1602A/C1669G/C1670G). This work was funded by a grant from the British Heart Foundation (PG/09/038/27320) (J.T.B.C., D.A.L., and R.d.G.).
Authorship
Contribution: R.d.G. designed research, performed experiments, interpreted data, and wrote the paper; D.A.L. designed research; and J.T.B.C. designed research, performed experiments, interpreted data, and wrote the paper
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rens de Groot, Centre for Haematology, Imperial College London, Fifth Floor Commonwealth Building, Hammersmith Hospital Campus, Du Cane Rd, W12 0NN, London, United Kingdom; e-mail: r.degroot@imperial.ac.uk.