Key Points
NF-κB differentially regulates CXCR4 expression on naïve and pathogenic CD8+ T cells.
CXCR4 expression on pathogenic T cells facilitates their trafficking to the BM in a mouse model of AA.
Abstract
Aplastic anemia (AA) is a disease characterized by T-cell–mediated destruction of bone marrow (BM) hematopoietic stem and progenitor cells. Physiologically, T cells migrate to the BM in response to chemokines, such as SDF-1α, the ligand for CXCR4. However, how T cells traffic to the BM in AA is poorly understood. CXCR4 is aberrantly expressed in immune-mediated diseases and its regulation by nuclear factor-κB (NF-κB) in cancer models is well documented. In this study, we show that CXCR4 is highly expressed on BM-infiltrating CD4+ and CD8+ T cells in a mouse model of AA. Inhibiting CXCR4 in AA mice, using CXCR4−/− splenocytes or AMD3100, significantly reduced BM infiltration of T cells. We also report that NF-κB occupancy at the CXCR4 promoter is enhanced in BM-infiltrating CD8+ T cells of AA mice. Moreover, inhibiting NF-κB signaling in AA mice using Bay11 or dehydroxymethylepoxyquinomicin, or transferring p50−/− splenocytes, decreased CXCR4 expression on CD8+ T cells, significantly reduced BM infiltration of T cells, and strongly attenuated disease symptoms. Remarkably, therapeutic administration of Bay11 significantly extended survival of AA mice. Overall, we demonstrate that CXCR4 mediates migration of pathogenic T cells to the BM in AA mice, and inhibiting NF-κB signaling may represent a novel therapeutic approach to treating AA.
Introduction
Aplastic anemia (AA) is a rare bone marrow failure (BMF) disease characterized by peripheral pancytopenia and hypoplastic bone marrow (BM).1 Most cases of acquired AA are idiopathic occurring both in children and adults, with roughly equal frequency in both genders.1,2 Studies of AA patients and animal models of BMF suggest acquired AA is an immune-mediated disease.3,4 Aberrant responses mediated by T helper type-1 (Th1), Th17, and cytotoxic CD8+ T cells, together with impaired function of regulatory T cells,5-10 culminate in BM destruction. Although the pathophysiology of AA is well defined, the molecular mechanisms responsible for T-cell infiltration into the BM during AA progression are poorly understood.
Small populations of mature CD4+ and CD8+ T cells reside in the BM. It is a priming site for antigen-specific T cells,11-13 as well as a homing site for memory T cells.14-16 Physiologically, T cells migrate to the BM in response to chemokines, such as stromal-cell derived factor-1α (SDF-1α) which is highly expressed by BM stromal cells.17,18 SDF-1α, also known as CXCL12, is the natural ligand for the chemokine receptor, CXCR4.19 SDF-1α–CXCR4 interactions initiate multiple signaling pathways that augment T cell co-stimulation, proliferation, cytokine production, migration, and survival.20-25 In T cells, activation through the T-cell receptor, polyclonal stimulation, SDF-1α interaction, and IFN-γ are stimuli that downregulate CXCR4, whereas signaling through IL-2, IL-4, IL-7, and IL-15 upregulates its expression.26-31
The nuclear factor-κB (NF-κB) family of transcription factors consists of five subunits, RelA (p65), RelB, c-Rel, NF-κB1 (p50), and NF-κB2 (p52), that function as homo- or heterodimers. NF-κB signaling plays a central role in T-cell activation, proliferation, differentiation, and survival.32
Dysregulated CXCR4 and NF-κB signaling pathways contribute to disease pathology in multiple immune-mediated diseases including multiple sclerosis, systemic lupus erythematosus, rheumatoid arthritis, and type 1 diabetes.33-41 Both signaling pathways have also been associated with hematopoietic and nonhematopoietic malignancies.42-44 Moreover, NF-κB–mediated regulation of CXCR4 expression and function in breast, pancreatic, gastric, prostatic, and ovarian cancers is well documented.45-51 However, the contribution of CXCR4 and NF-κB signaling pathways to the pathology of acquired AA has not previously been explored.
Through pharmacologic and genetic approaches, we demonstrate that CXCR4 mediates migration of pathogenic T cells to the BM in an established mouse model of immune-mediated AA.5 We further show that CXCR4 is differentially regulated by NF-κB in naïve and BM-infiltrating CD8+ T cells. Inhibiting NF-κB signaling in AA mice decreased CXCR4 expression on BM-infiltrating CD8+ T cells, significantly reduced BM infiltration of T cells, and strongly attenuated disease symptoms. Finally, we show that therapeutic inhibition of NF-κB signaling significantly prolonged the survival of AA mice.
Materials and methods
Animals
Animal studies were conducted in compliance with the Institutional Animal Care and Use Committee of the University of Massachusetts Amherst. F1 progeny were obtained by crossing BALB/c females with C57BL/6 males. Conditional Cxcr4 knockout (CXCR4−/−) mice were generated on a C57BL/6J background by crossing Cxcr4fl/fl mice (B6.129P2-Cxcr4tm2Yzo/J) to Mx1-cre+/− mice (B6.Cg-Tg[Mx1-cre]1Cgn/J). Cxcr4fl/flMx1-cre+/− (CXCR4−/−), Cxcr4fl/flMx1-cre−/−, and Mx1-cre+/− mice were administered polyI:polyC (12 to 15 μg/g body weight; Imgenex, San Diego, CA) via IP injection every other day for 5 days. Mice were rested for 3 weeks and then used as a source of donor splenocytes. p50 knockout (B6.Cg-Nfkb1tm1Bal/J; p50−/−), BALB/c, C57BL/6, Cxcr4fl/fl, and Mx1-cre+/− parental strains were obtained from the Jackson Laboratory (Bar Harbor, ME). Mice between 7 to 12 weeks were used in experiments.
BMF induction and analyses
F1 progeny were irradiated (3Gy,137Cs source); 4 to 6 hours later, BMF was induced with 5 × 107 splenocytes (IP injection) from age- and gender-matched C57BL/6 (wild-type [WT] or knockout) donors.5 Mice were harvested on day +17. For survival studies, mice were humanely euthanized when they could no longer eat or drink. BM cells were flushed from tibias and femurs of legs using 5% fetal bovine serum (FBS)/phosphate-buffered saline (PBS). Splenocytes were passed through a 40 μM filter, red blood cells (RBCs) lysed with ACK buffer, and white blood cells (WBCs) enumerated using trypan blue exclusion. Circulating WBCs and RBCs were counted using a HemaTrue Hematology Analyzer (Heska). For quantitative real-time polymerase chain reaction (qRT-PCR) studies, T cells were isolated from spleens or BM of AA mice using anti-mouse CD4 and CD8 magnetic particles (BD Biosciences) and separated using the BD IMag system.
In vivo administration of CXCR4 and NF-κB inhibitors
For CXCR4 inhibition studies, mice were treated on day +7 postdisease induction with AMD3100 (5 mg/kg per day; Calbiochem) administered via IP injection continuing until day +16. Control mice received equivalent volumes of PBS. For NF-κB inhibition studies, mice were administered dehydroxymethylepoxyquinomicin (DHMEQ) (30 mg/kg per day; provided by A. Fauq) or Bay11-7085 (Bay11; 5 mg/kg every other day; Calbiochem) via IP injection, beginning 1 hour after disease induction and continuing until day +16 postdisease induction. Control mice received equivalent volumes of dimethylsulfoxide (DMSO). For survival studies, mice were given Bay11 via IP injection (5 mg/kg every other day) from days +7 to +17 postdisease induction, at which time treatment was discontinued. Control mice received equivalent volumes of DMSO.
Histology
On day +17 postdisease induction, sterna were harvested, fixed overnight in 10% neutral buffered formalin (VWR), decalcified for 48 hours in Cal-Rite (Richard-Allan Scientific), then preserved in 70% ethanol at 4°C until processed, paraffin-embedded, sectioned, and stained with hematoxylin and eosin.
Mixed lymphocyte reaction and chemotaxis assay
To generate BM-derived dendritic cells (BMDCs), BM cells from F1 progeny were cultured (106 cells/mL) in RPMI 1640 medium supplemented with 10% FBS (Gibco), 2 mM l-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, and 20 ng/mL granulocyte macrophage colony-stimulating factor (R&D Systems), and incubated at 37°C with 5% CO2. On days 2 and 4 postculture, half the media was removed and replaced with fresh supplemented media. On day 6 postculture, nonadherent cells were harvested and cultured in fresh supplemented media for 2 additional days. For mixed leukocyte reaction, BMDCs were cocultured with bulk splenocytes (1:10 from age- and gender-matched C57BL/6 mice) in a 1:1 mixture of RPMI 1640 and Dulbecco’s modified Eagle medium supplemented with 10% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, 100 U/mL penicillin, and 100 μg/mL streptomycin in 96-well round-bottom plates and incubated at 37°C with 7% CO2 for 12 days. To inhibit NF-κB, 1 μM Bay11 or an equivalent volume of DMSO was added at time of plating. From days 6 to 12 postculture, nonadherent cells were harvested every other day, and CXCR4 expression on CD4+ and CD8+ T cells was analyzed by flow cytometry. On day 8 postculture, nonadherent cells were harvested to evaluate their chemotactic response to SDF-1α. Chemotaxis assays were performed in 24-well plate Transwell inserts (5 μm polycarbonate membrane, 6.5 mm insert; Corning Costar); 600 μl of RPMI with 10% FBS with or without 100 ng/mL of SDF-1α (R&D Systems) was added to wells and 0.5 × 106 cells in 0.1 μl of RPMI with 10% FBS were added to inserts, and incubated at 37°C with 5% CO2. After 3 hours, inserts were removed and migrated cells harvested from wells. CD4+ and CD8+ T cells were enumerated by flow cytometry. Results are expressed as chemotactic index (number of cells migrated into 100 ng/mL SDF-1α wells divided by number of cells migrated into 0 ng/mL SDF-1α wells). Media and supplements were from Lonza Group, unless otherwise specified.
Flow cytometry
Single-cell suspensions from spleens and BM were surface-stained with PerCP-conjugated anti-CD4 (RM4-5; BD Pharmingen), PE-Cy7-conjugated anti-CD8a (53-6.7; eBioscience), APC-conjugated anti-CXCR4 (2B11; eBioscience), APC-conjugated anti-CXCR7 (11G8; R&D Systems), APC-conjugated anti-CX3CR1 (polyclonal; R&D Systems), PE-conjugated anti-CCR5 (CTC5; R&D Systems), and PE-conjugated anti-CXCR3 (220803; R&D Systems). Samples were acquired on an LSRII flow cytometer and analyzed using FACSDiva acquisition software (Becton Dickinson). Analyses of fluorescence-activated cell sorter data were performed using FACSDiva or FloJo software (Tree Star, Ashland, OR).
Cytometric bead array
Plasma cytokine levels were determined using Th1/Th2 cytometric bead array kits (BD Biosciences) following the manufacturer’s protocol. Data were acquired on an LSRII flow cytometer and analyzed using FCAP array software (BD Biosciences).
RNA isolation and qRT-PCR
Total RNA was extracted using RNAqueous Kits (Ambion) according to the manufacturer’s protocol. RNA (1 μg) was reverse transcribed to complementary DNA using dNTPs (Roche), M-MuLV reverse transcriptase reaction buffer (New England Biolabs, Inc.), oligo-(dT)12–18 (Invitrogen), RNase inhibitor (Promega), and M-MuLV reverse transcriptase (New England Biolabs, Inc.) on a Mastercycler Gradient Thermal Cycler (Eppendorf). qRT-PCR was performed in duplicate with SYBR Premix Ex Taq (Takara Bio, Inc.) on a Stratagene Mx3000P quantitative PCR system (Agilent Technologies). Primer sequences were as follows: Cxcr4: forward, 5′-GAC TGG CAT AGT CGG CAA TG-3′; reverse, 5′-AGA AGG GGA GTG TGA TGA CAA A-3′; and Actb: forward, 5′-GGC TGT ATT CCC CTC CAT CG-3′; reverse, 5′-CCA GTT GGT AAC AAT GCC ATG T-3′. qRT-PCR conditions were as follows: 95°C for 1 minute, 95°C for 25 seconds, 62°C for 25 seconds (35 cycles), 95°C for 1 minute, 62°C for 1 minute, and 95°C for 30 seconds. Relative expression of Cxcr4 was determined using the 2−ΔΔCt method. Results are reported as fold-change in gene expression, normalized to Actb and relative to irradiation controls.
ChIP
Chromatin immunoprecipitation (ChIP) was performed as described.5 Briefly, cells were crosslinked with 1% formaldehyde, lysed in sodium dodecyl sulfate (SDS) lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris, pH8.1), and sonicated with a Bioruptor Sonicator (Diagenode). Cell lysates were precleared overnight with 5 μg anti–c-Rel (clone C), anti–NF-κB p50 (clone C-19), normal rabbit IgG, or normal goat IgG at 4°C (all from Santa Cruz Biotechnology, Inc.). Protein–DNA complexes were recovered with ChIP-grade protein G agarose beads, washed, eluted with elution buffer (1% SDS, 0.1 M NaHCO3), and reverse crosslinked overnight at 65°C. DNA was purified by proteinase K digestion and extracted with QIAEX II gel extraction kit (QIAGEN). Two regions of the Cxcr4 promoter containing putative NF-κB p50 and c-Rel binding sites were amplified by PCR. Primer sets were as follows: primer set I (product size 185 bp) forward, 5′-GGC TGA CCT CCT CTT TGT CAT C-3′; reverse, 5′-TGT TGG TGG CGT GGA CAA TA-3′; and primer set II (product size 331 bp) forward, 5′-CAT CAG TCA GGG GGA TGA CA-3′; reverse, 5′-GAT GGA GAT CCA CTT GTG CA-3′. PCR conditions were 95°C for 2 minutes, 95°C for 30 seconds, 56°C for 30 seconds, 72°C for 1 minute (38 cycles), and 72°C for 5 minutes.
Statistical analysis
Results are the mean ± SEM. Statistical analyses were performed using GraphPad Prism 5.0 (GraphPad Software). P values were calculated using an unpaired two-tailed Student t test, one- or two-way analysis of variance (ANOVA) with post-tests as indicated. Survival curves were generated using the Kaplan–Meier method and survival differences were determined with a Mantel-Cox log-rank test. P values ≤ .05 were considered statistically significant.
Results
CXCR4 is highly expressed on T cells of AA mice
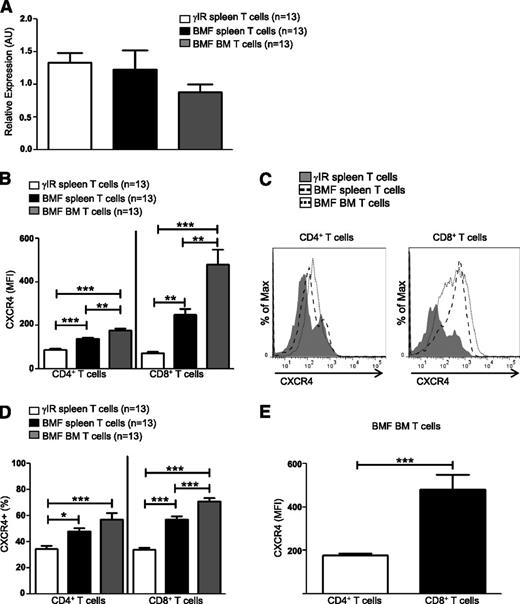
T cells use CXCR4–SDF-1α interactions to traffic to the BM during homeostatic processes,17,18 but how pathogenic T cells are targeted to the BM during AA is not well understood. Here, we sought to determine whether CXCR4 contributes to the pathogenesis of immune-mediated AA. Using a well-characterized, lymphocyte transfer mouse model of AA,5 we first assessed transcript and protein expression of CXCR4. Seventeen days postdisease induction, Cxcr4 transcripts in T cells from spleens and BM were expressed at similar levels both in AA mice and irradiation controls (Figure 1A); however, for many genes, there is only a modest correlation between messenger RNA levels and their translated proteins.52 When we further assessed protein levels, we found that the surface expression of CXCR4 was significantly higher on T cells isolated from spleens and BM of AA mice (Figure 1B-C), as were percentages of CXCR4+ T cells (Figure 1D), suggesting that posttranscriptional regulation of CXCR4 differs between control and pathogenic T cells. We further noted that CXCR4 was expressed more highly on a per cell basis on BM-infiltrating CD8+ T cells, compared with CD4+ T cells (Figure 1E).
CXCR4 is highly expressed on T cells of AA mice. F1 hybrid mice were irradiated only (γIR control) or AA was induced with 5 × 107 WT C57BL/6 splenocytes. Mice were sacrificed on day +17 postdisease-induction and T cells were isolated. (A) Relative expression of Cxcr4 transcript levels in T cells from spleens (BMF spleen) and BM (BMF BM) of AA mice were determined by quantitative PCR and compared with expression in T cells of spleens (γIR spleen) from irradiation controls; n = 13 mice per group. (B) Median fluorescence intensity (MFI), a measure of CXCR4 protein expression, on spleen and BM T cells of AA mice was determined by flow cytometry and compared with expression on spleen T cells of irradiated controls; n = 13 mice per group. (C) Representative comparative histograms of CXCR4 MFI on T cells from spleens and BM from (B). (D) Percent CXCR4 positive T cells on T cells from spleens and BM of AA mice, compared with T cells isolated from spleens of irradiation controls; n = 13 mice per group. (E) Comparison of CXCR4 expression on BM-infiltrating CD4+ and CD8+ T cells of AA mice; n = 13 mice per group. Data represent the mean ± SEM, and were analyzed by one-way ANOVA plus Tukey post-test (A-B,D), or two-tailed unpaired Student t test (E). *P < .05; **P < .01; ***P < .001.
CXCR4 is highly expressed on T cells of AA mice. F1 hybrid mice were irradiated only (γIR control) or AA was induced with 5 × 107 WT C57BL/6 splenocytes. Mice were sacrificed on day +17 postdisease-induction and T cells were isolated. (A) Relative expression of Cxcr4 transcript levels in T cells from spleens (BMF spleen) and BM (BMF BM) of AA mice were determined by quantitative PCR and compared with expression in T cells of spleens (γIR spleen) from irradiation controls; n = 13 mice per group. (B) Median fluorescence intensity (MFI), a measure of CXCR4 protein expression, on spleen and BM T cells of AA mice was determined by flow cytometry and compared with expression on spleen T cells of irradiated controls; n = 13 mice per group. (C) Representative comparative histograms of CXCR4 MFI on T cells from spleens and BM from (B). (D) Percent CXCR4 positive T cells on T cells from spleens and BM of AA mice, compared with T cells isolated from spleens of irradiation controls; n = 13 mice per group. (E) Comparison of CXCR4 expression on BM-infiltrating CD4+ and CD8+ T cells of AA mice; n = 13 mice per group. Data represent the mean ± SEM, and were analyzed by one-way ANOVA plus Tukey post-test (A-B,D), or two-tailed unpaired Student t test (E). *P < .05; **P < .01; ***P < .001.
Inhibiting CXCR4 in AA mice reduces T-cell homing to the BM
The natural ligand for CXCR4, SDF-1α, is present in the BM at high concentrations.53 We hypothesized that sustained CXCR4 expression on activated T cells might aid in their trafficking to the BM in diseased animals. Therefore, we asked whether reducing the CXCR4 expression or preventing SDF-1α–CXCR4 interactions would affect BM infiltration of T cells in a mouse model of AA.
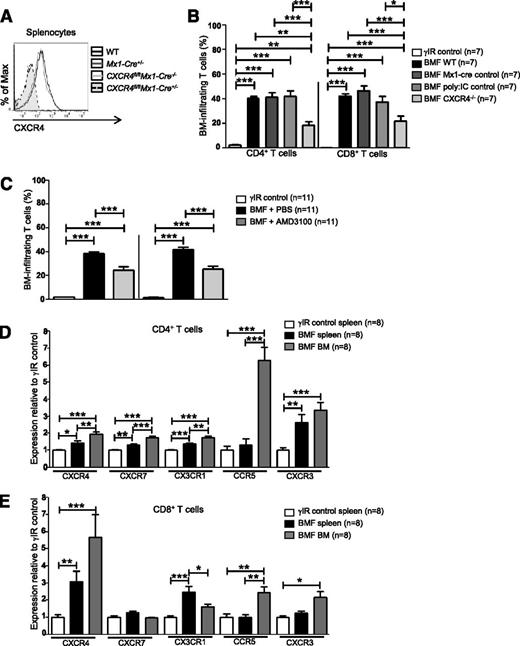
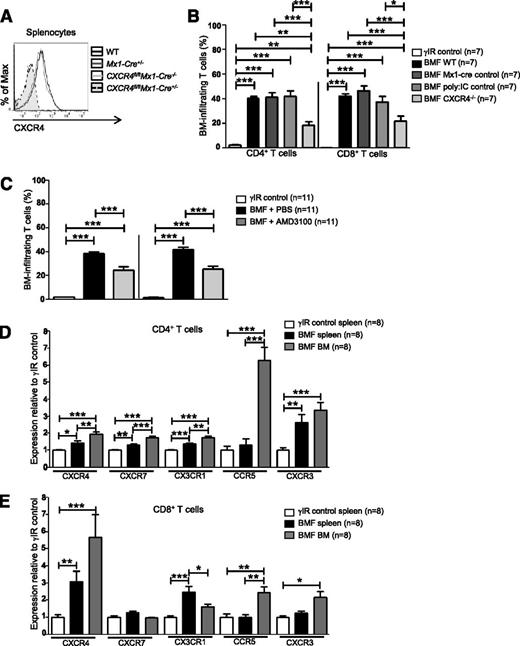
Cxcr4 knockout mice are embryonic lethal due to defects in B-cell lymphopoiesis and myelopoiesis, heart and cerebellar development, and vascularization.54,55 Therefore, we used Cre/loxP DNA recombination to conditionally delete Cxcr4 from splenocytes of Cxcr4fl/flMx1-cre+/− (CXCR4−/−) donor mice via serial injections of polyinosinic:polycytidylic acid (poly:IC) (Figure 2A).56 AA mice receiving CXCR4−/− splenocytes had significantly fewer BM-infiltrating CD4+ and CD8+ T cells compared with mice who received control splenocytes (Cxcr4fl/flMx1- cre−/−, Mx1-cre+/−, or WT splenocytes; Figure 2B).
Inhibiting CXCR4 in AA mice reduces T-cell homing to the BM. (A) Representative histograms of CXCR4 expression on splenocytes from WT C57BL/6 mice, poly:IC-treated Mx1-cre+/− mice (Mx1-cre+/−; Mx1-cre only control), poly:IC-treated CXCR4fl/flMx1-cre−/− mice (CXCR4fl/flMx1-cre−/−; poly:IC control), or poly:IC-treated CXCR4fl/flMx1-cre+/− mice (CXCR4fl/flMx1-cre+/−; CXCR4−/−). (B) Donor splenocytes from (A) were used to induce AA in F1 recipient mice (BMF). On day +17 postdisease induction, percentages of BM-infiltrating CD4+ and CD8+ T cells in AA mice were assessed by flow cytometry and compared with mice that were irradiated only (γIR control); n = 7 mice per group. (C) Mice were induced with AA (BMF), and from days +7 to +16 postdisease induction, were treated with PBS (BMF + PBS) or with the CXCR4 antagonist, AMD3100 (BMF + AMD3100). Percentages of BM-infiltrating CD4+ and CD8+ T cells in AA mice were determined as in (B); n = 11 mice per group. (D-E) F1 hybrid mice were irradiated only (γIR control) or AA was induced by transferrin 5 × 107 WT C57BL/6 splenocytes (BMF). On day +17 postdisease induction, the MFI of CXCR4, CXCR7, CX3CR1, CCR5, and CXCR3 on (D) CD4+ and (E) CD8+ T cells from the spleens and BM of AA mice or irradiation controls was assessed by flow cytometry. Expression was relative to γIR control and was calculated by dividing the MFI of BMF samples by the mean MFI of γIR controls; n = 8 mice per group. Data are the mean ± SEM, and were analyzed by one-way ANOVA plus Tukey post-test. *P < .05; **P < .01; ***P < .001.
Inhibiting CXCR4 in AA mice reduces T-cell homing to the BM. (A) Representative histograms of CXCR4 expression on splenocytes from WT C57BL/6 mice, poly:IC-treated Mx1-cre+/− mice (Mx1-cre+/−; Mx1-cre only control), poly:IC-treated CXCR4fl/flMx1-cre−/− mice (CXCR4fl/flMx1-cre−/−; poly:IC control), or poly:IC-treated CXCR4fl/flMx1-cre+/− mice (CXCR4fl/flMx1-cre+/−; CXCR4−/−). (B) Donor splenocytes from (A) were used to induce AA in F1 recipient mice (BMF). On day +17 postdisease induction, percentages of BM-infiltrating CD4+ and CD8+ T cells in AA mice were assessed by flow cytometry and compared with mice that were irradiated only (γIR control); n = 7 mice per group. (C) Mice were induced with AA (BMF), and from days +7 to +16 postdisease induction, were treated with PBS (BMF + PBS) or with the CXCR4 antagonist, AMD3100 (BMF + AMD3100). Percentages of BM-infiltrating CD4+ and CD8+ T cells in AA mice were determined as in (B); n = 11 mice per group. (D-E) F1 hybrid mice were irradiated only (γIR control) or AA was induced by transferrin 5 × 107 WT C57BL/6 splenocytes (BMF). On day +17 postdisease induction, the MFI of CXCR4, CXCR7, CX3CR1, CCR5, and CXCR3 on (D) CD4+ and (E) CD8+ T cells from the spleens and BM of AA mice or irradiation controls was assessed by flow cytometry. Expression was relative to γIR control and was calculated by dividing the MFI of BMF samples by the mean MFI of γIR controls; n = 8 mice per group. Data are the mean ± SEM, and were analyzed by one-way ANOVA plus Tukey post-test. *P < .05; **P < .01; ***P < .001.
We next abrogated SDF-1α–CXCR4 binding using the highly selective CXCR4 antagonist, AMD3100.57 To prevent SDF-1α–CXCR4 interactions during T-cell expansion, AA mice were given AMD3100 (5 mg/kg per day) for 10 days, beginning day +7 postdisease induction. On day +17, we assessed T-cell accumulation in the BM and noted a significant, albeit incomplete reduction, in BM-infiltrating CD4+ and CD8+ T cells in AA mice treated with AMD3100, compared with those treated with vehicle (Figure 2C). Therefore, we analyzed the expression of additional chemokine receptors associated with BM trafficking. Compared with irradiated controls, T cells from spleens and BM of AA mice showed modestly increased CXCR7, CX3CR1, CCR5, and CXCR3 (Figure 2D-E). Among these, CCR5 showed the greatest relative increase on BM CD4+ T cells, whereas CXCR4 was the most highly upregulated on CD8+ T cells (Figure 2D-E). Taken together, these data provide evidence that in AA mice, pathogenic CD8+ T cells aberrantly express and use CXCR4 to traffic to the BM, although additional chemokine receptors may also contribute to this process.
Inhibiting NF-κB signaling reduces CXCR4 expression and abrogates T-cell migration
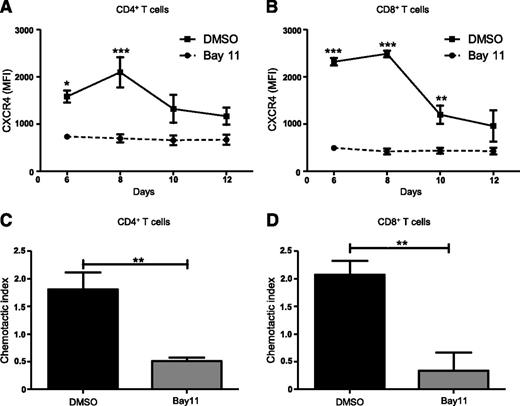
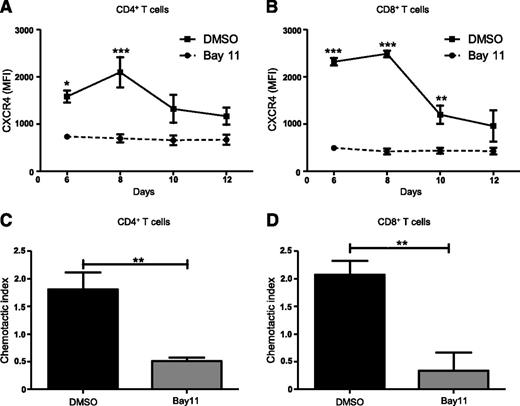
NF-κB signaling is central to the activation, proliferation, differentiation, and survival of T cells.32 NF-κB regulates CXCR4 expression and subsequent metastasis in a range of malignancies including breast, pancreatic, gastric, prostatic, and ovarian cancers.45-51 However, NF-κB–mediated regulation of CXCR4 expression on T cells has not been previously investigated. Therefore, we asked whether blocking NF-κB signaling would modulate CXCR4 expression on T cells. We used a mixed lymphocyte reaction in which C57BL/6 splenocytes treated with the NF-κB inhibitor, Bay11 (1 μM), or vehicle only, were cocultured with BMDCs from F1 progeny (C57BL / 6×BALB/c), and then analyzed T cells for CXCR4 expression using flow cytometry. Compared with controls, Bay11-treated CD4+ T cells showed significantly diminished CXCR4 expression after 6 and 8 days of culture (Figure 3A), whereas Bay11-treated CD8+ T cells displayed diminished CXCR4 levels on days 6, 8, and 10 of culture (Figure 3B). This is in contrast to DMSO-treated CD4+ and CD8+ T cells, which display elevated CXCR4 expression. We next determined how abrogating NF-κB signaling affects T-cell migration in response to SDF-1α. Treating CD4+ or CD8+ T cells with Bay11 significantly impaired their ability to migrate toward SDF-1α, compared with control cells treated with vehicle only (Figure 3C-D). These findings support the notion that NF-κB signaling regulates CXCR4 expression on CD4+ and CD8+ T cells, which, in turn, mediates their migration across an SDF-1α gradient.
Inhibiting NF-κB signaling reduces CXCR4 expression and abrogates T-cell migration. Splenocytes from C57BL/6 mice were treated with DMSO or Bay11 (1 μM) and cocultured for 12 days with BMDCs from F1 hybrids. CXCR4 expression was assessed by flow cytometry on (A) CD4+ and (B) CD8+ T cells; n = 3. On day 8 postculture, nonadherent cells were harvested from cocultures (as above) and their chemotactic response to SDF-1α was analyzed. Cells were added to Transwell inserts; 0 or 100 ng/ml SDF-1α was added to wells. Cultures were incubated for 3 hours, at which time migrated cells were harvested from wells and CD4+ and CD8+ T cells were enumerated by flow cytometry. The chemotactic index (number of cells migrated into 100 ng/ml SDF-1α wells/number of cells migrated into 0 ng/ml SDF-1α wells) was calculated for DMSO- or Bay11-treated (C) CD4+ and (D) CD8+ T cells; n = 3. Data are the mean ± SEM, and were analyzed using two-way ANOVA plus Bonferroni post-test (A-B) or two-tailed unpaired Student t test (C-D). *P < .05; **P < .01; ***P < .001.
Inhibiting NF-κB signaling reduces CXCR4 expression and abrogates T-cell migration. Splenocytes from C57BL/6 mice were treated with DMSO or Bay11 (1 μM) and cocultured for 12 days with BMDCs from F1 hybrids. CXCR4 expression was assessed by flow cytometry on (A) CD4+ and (B) CD8+ T cells; n = 3. On day 8 postculture, nonadherent cells were harvested from cocultures (as above) and their chemotactic response to SDF-1α was analyzed. Cells were added to Transwell inserts; 0 or 100 ng/ml SDF-1α was added to wells. Cultures were incubated for 3 hours, at which time migrated cells were harvested from wells and CD4+ and CD8+ T cells were enumerated by flow cytometry. The chemotactic index (number of cells migrated into 100 ng/ml SDF-1α wells/number of cells migrated into 0 ng/ml SDF-1α wells) was calculated for DMSO- or Bay11-treated (C) CD4+ and (D) CD8+ T cells; n = 3. Data are the mean ± SEM, and were analyzed using two-way ANOVA plus Bonferroni post-test (A-B) or two-tailed unpaired Student t test (C-D). *P < .05; **P < .01; ***P < .001.
The NF-κB p50 subunit differentially binds the Cxcr4 promoter in T cells of AA mice
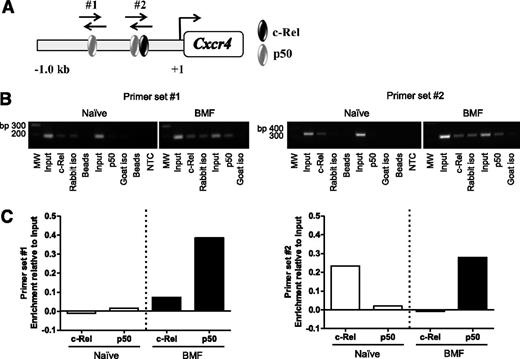
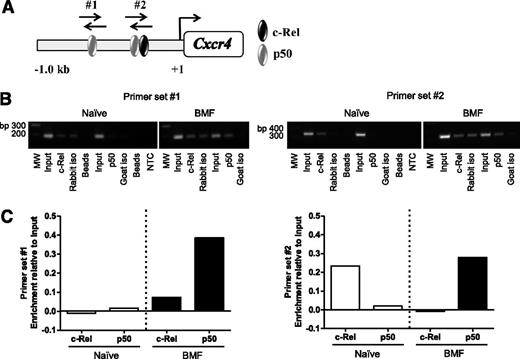
Previous reports using human cancer cell lines confirmed NF-κB binding sites in the CXCR4 promoter; however, Cxcr4 regulation in T cells is ill-defined.45,47-50 Therefore, we asked whether NF-κB directly regulates Cxcr4 in BM-infiltrating T cells from AA mice. We analyzed a 1 Kb region upstream of the transcriptional start site in the Cxcr4 promoter and identified 2 putative binding sites for NF-κB subunits, c-Rel and p50 (Figure 4A). Using chromatin derived from naïve CD8+ T cells or from BM-infiltrating CD8+ T cells isolated from AA mice, we performed ChIP with antibodies specific for c-Rel and p50. Unlike in naïve cells, we detected increased p50 occupancy at both sites in the Cxcr4 promoter in pathogenic CD8+ T cells. We also observed differential binding of c-Rel in naïve and BM-infiltrating CD8+ T cells (Figure 4B-C). Collectively, these data reveal that NF-κB–binding to the Cxcr4 promoter is increased in BM-infiltrating CD8+ T cells from AA mice. Furthermore, differences in NF-κB occupancy at the regions we investigated suggest that NF-κB differentially regulates Cxcr4 in naïve and pathogenic CD8+ T cells.
The NF-κB p50 subunit differentially binds the Cxcr4 promoter in T cells of AA mice. (A) Schematic representation of the Cxcr4 promoter showing relative location of predicted binding sites for NF-κB subunits, p50 and c-Rel, and regions amplified by primer sets #1 and #2 (not shown to scale). (B) Representative image of agarose gel showing 2 amplified regions of the Cxcr4 promoter. Chromatin was from BM-infiltrating CD8+ cells of mice induced with AA (BMF; right) or spleen-infiltrating CD8+ T cells of noninduced mice (Naïve; left) immunoprecipitated using antibodies specific for p50 and c-Rel. (C) Quantification of band intensities of Naïve and BMF samples subjected to ChIP in (B). Data are representative of 2 independent experiments.
The NF-κB p50 subunit differentially binds the Cxcr4 promoter in T cells of AA mice. (A) Schematic representation of the Cxcr4 promoter showing relative location of predicted binding sites for NF-κB subunits, p50 and c-Rel, and regions amplified by primer sets #1 and #2 (not shown to scale). (B) Representative image of agarose gel showing 2 amplified regions of the Cxcr4 promoter. Chromatin was from BM-infiltrating CD8+ cells of mice induced with AA (BMF; right) or spleen-infiltrating CD8+ T cells of noninduced mice (Naïve; left) immunoprecipitated using antibodies specific for p50 and c-Rel. (C) Quantification of band intensities of Naïve and BMF samples subjected to ChIP in (B). Data are representative of 2 independent experiments.
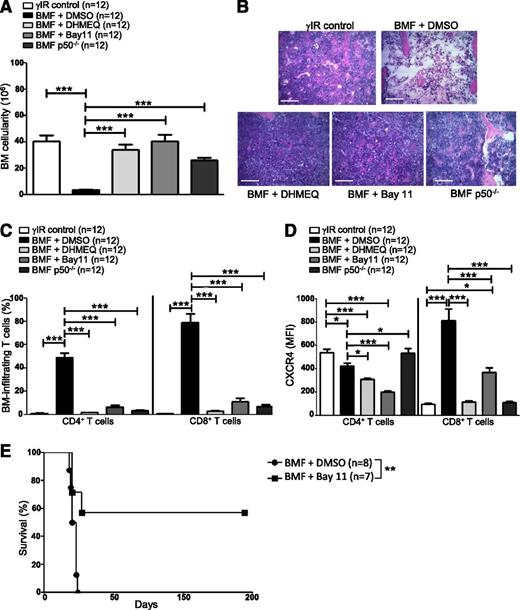
Inhibiting NF-κB signaling in AA mice reduces CXCR4 expression in T cells and attenuates AA
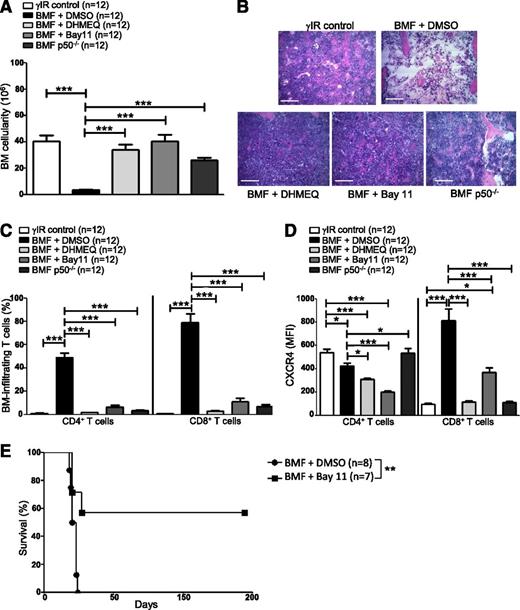
Blocking NF-κB signaling impairs bone metastasis in experimental breast cancer models by preventing CXCR4 expression.58 We inhibited NF-κB signaling in AA mice pharmacologically, using DHMEQ or Bay11, or by transferring p50−/− splenocytes, to determine what effect this would have on T-cell infiltration to the BM and disease progression overall. AA mice were treated with DHMEQ (30 mg/kg per day) or Bay11 (5 mg/kg every other day) beginning 1 hour after disease induction and continuing for 16 days. On day +17 postdisease induction, mice were humanely euthanized to assess disease severity. Inhibiting NF-κB activity in AA mice robustly protected BM cellularity (Figure 5A-B), and this protection extended to circulating WBCs and RBCs (see supplemental Figure 1A-B, available on the Blood Web site). BM-infiltrating CD4+ and CD8+ T cells (Figure 5C), and circulating IFN-γ and tumor necrosis factor (supplemental Figure 1C-D) were negligible in AA mice with impaired NF-κB signaling, compared with vehicle-treated controls.
Inhibiting NF-κB signaling in AA mice reduces CXCR4 expression in T cells and attenuates AA. F1 hybrid mice were irradiated only (γIR control) or AA was induced with 5 × 107 WT C57BL/6 splenocytes. Mice were treated with DMSO (BMF + DMSO), DHMEQ (BMF + DHMEQ), or Bay11 (BMF + Bay11). In some mice, AA was induced with 5 × 107 p50−/− splenocytes (BMF p50−/−). On day +17 postdisease induction, mice were harvested and (A) total BM cellularity was determined by trypan blue exclusion; n = 12 mice per group. (B) Representative hematoxylin and eosin staining of sternum from 1 representative animal each of irradiation control (γIR control), DMSO-treated AA mouse (BMF + DMSO), DHMEQ-treated AA mouse (BMF + DHMEQ), Bay11-treated AA mouse (BMF + Bay11), and 1 mouse that received p50−/− splenocytes (BMF p50−/−). Scale bar = 200 μM. (C) Percentages of BM-infiltrating CD4+ and CD8+ T cells in irradiation controls or AA mice that received DMSO, DHMEQ, Bay11, or p50−/− splenocytes were determined by flow cytometry; n = 12 mice per group. (D) MFI of CXCR4 protein expression on BM-infiltrating CD4+ and CD8+ T cells of AA mice that received DMSO, DHMEQ, Bay11, or p50−/− splenocytes was assessed by flow cytometry and compared with CXCR4 MFI on BM-infiltrating T cells of irradiation controls; n = 12. (E) Kaplan–Meier survival estimates of AA mice whose disease was induced with WT splenocytes and treated with DMSO (n = 8) or Bay11 (n = 7), beginning on day +7 postdisease induction and continuing until day +17 postdisease induction. Data are the mean ± SEM, and analyzed using one-way ANOVA plus Tukey post-test or log-rank test for survival estimates. *P < .05; **P < .01; ***P < .001.
Inhibiting NF-κB signaling in AA mice reduces CXCR4 expression in T cells and attenuates AA. F1 hybrid mice were irradiated only (γIR control) or AA was induced with 5 × 107 WT C57BL/6 splenocytes. Mice were treated with DMSO (BMF + DMSO), DHMEQ (BMF + DHMEQ), or Bay11 (BMF + Bay11). In some mice, AA was induced with 5 × 107 p50−/− splenocytes (BMF p50−/−). On day +17 postdisease induction, mice were harvested and (A) total BM cellularity was determined by trypan blue exclusion; n = 12 mice per group. (B) Representative hematoxylin and eosin staining of sternum from 1 representative animal each of irradiation control (γIR control), DMSO-treated AA mouse (BMF + DMSO), DHMEQ-treated AA mouse (BMF + DHMEQ), Bay11-treated AA mouse (BMF + Bay11), and 1 mouse that received p50−/− splenocytes (BMF p50−/−). Scale bar = 200 μM. (C) Percentages of BM-infiltrating CD4+ and CD8+ T cells in irradiation controls or AA mice that received DMSO, DHMEQ, Bay11, or p50−/− splenocytes were determined by flow cytometry; n = 12 mice per group. (D) MFI of CXCR4 protein expression on BM-infiltrating CD4+ and CD8+ T cells of AA mice that received DMSO, DHMEQ, Bay11, or p50−/− splenocytes was assessed by flow cytometry and compared with CXCR4 MFI on BM-infiltrating T cells of irradiation controls; n = 12. (E) Kaplan–Meier survival estimates of AA mice whose disease was induced with WT splenocytes and treated with DMSO (n = 8) or Bay11 (n = 7), beginning on day +7 postdisease induction and continuing until day +17 postdisease induction. Data are the mean ± SEM, and analyzed using one-way ANOVA plus Tukey post-test or log-rank test for survival estimates. *P < .05; **P < .01; ***P < .001.
Moreover, when AA mice received NF-κB inhibitors or p50−/− splenocytes, a significantly lower percentage of BM CD8+ T cells expressed CXCR4 (supplemental Figure 1E), and the per cell expression of CXCR4 on BM CD8+ T cells was significantly lower compared with control-treated AA mice (Figure 5D). Consistent with the results of the ChIP (Figure 4), these data support the notion that NF-κB, and more specifically p50, play an important role in regulating CXCR4 expression in pathogenic CD8+ T cells.
Finally, we evaluated the survival benefit of pharmacologically limiting NF-κB signaling in AA mice under clinically relevant conditions. AA mice were treated with Bay11 (5 mg/kg every other day) or vehicle only (DMSO) from days +7 to +17 postdisease induction, at which time treatment was discontinued. Compared with DMSO-treated mice, for which the median survival time was 19.5 days (range, 18 to 24 days), Bay11 treatment significantly extended survival in AA mice, with 4 of 7 mice fully rescued from lethal BMF (P < .01) (Figure 5E). Collectively, our data demonstrate that attenuating NF-κB signaling in a mouse model of AA reduces CXCR4 expression on CD8+ T cells, prevents T-cell homing to the BM, and ameliorates disease symptoms.
Discussion
AA is primarily driven by an aberrant Th1 immune response, whereby activated T cells traffic to the BM and mediate destruction. Differential expression of chemokine receptors and adhesion molecules on Th-cell subsets endow these T cells with distinct migratory abilities and selective recruitment into peripheral tissues.27,59-62 The chemokine receptor, CXCR4, facilitates cellular chemotaxis in response to its natural ligand, SDF-1α, found in high concentrations in the BM.17,18,63-65 Aberrantly elevated CXCR4 has been reported in numerous autoimmune conditions, and is also thought to provide a means for certain cancerous cells to home to SDF-1α–enriched sites, such as the BM.33-39,47 CXCR4 is highly-expressed on resting T cells, including naïve and memory T cells, and is downregulated during T-cell activation.26,30,31,60 Using inhibitors and genetic approaches, we provide evidence that BM-infiltrating CD4+ and CD8+ T cells from AA mice show high CXCR4 expression, suggesting CXCR4–SDF-1α interaction may be one mechanism pathogenic T cells use to traffic to the BM during AA progression.
Chemokine receptors, including CX3CR1 and CCR5, facilitate migration of normal and malignant cells to the BM, whereas CXCR7 has been identified as an additional receptor for SDF-1α.66-69 Data presented in this study (Figure 2D-E) are in concordance with previous reports describing elevated CX3CR1 on peripheral and BM T cells, and increased expression of CX3CR1 and CXCR3 in BM T cells of patients with severe AA.67,70,71 Reports also indicate peripheral and BM CD4+ regulatory T cells from patients with severe AA have reduced CXCR4 expression, compared with those of healthy controls, with impaired ability to home to the BM and suppress pathogenic T cells.10 In AA mice, CCR5 expression on BM-infiltrating CD4+ T cells showed the greatest increase, whereas on BM CD8+ T cells, CXCR4 exhibited the highest relative gain. Residual BM T cells could be detected in AA mice whose disease was induced with CXCR4−/− splenocytes or were treated with AMD3100, indicating chemokine receptors other than CXCR4 may aid in T cell trafficking to the BM. Indeed, we observed modestly augmented CXCR7, CX3CR1, and CXCR3 on BM T cells and their expression levels remained unchanged when CXCR4 was inhibited (data not shown). Thus, dysregulated expression of chemokine receptors may be a general feature of AA progression in mice and in humans.
In breast cancer cell lines, NF-κB directly regulates Cxcr4 expression, which enhances motility in vitro and in vivo metastasis to the BM in animal models.47 Consistent with this notion, we observed increased p50 occupancy at two distinct NF-κB binding sites in the Cxcr4 promoter in BM-infiltrating CD8+ T cells. These sites were not similarly bound in naïve CD8+ T cells, suggesting that differential NF-κB binding to the Cxcr4 promoter in BM-infiltrating CD8+ T cells contributes to aberrant CXCR4 expression. Inhibitory or genetic approaches that limited NF-κB signaling decreased CXCR4 expression on CD8+ T cells, and this further correlated with their reduced trafficking to the BM. However, compared with approaches that only abrogated CXCR4–SDF-1α interactions in AA mice, limiting NF-κB activity resulted in a more profound reduction in T-cell infiltration to the BM and negligible circulating IFN-γ and tumor necrosis factor. Finally, inhibiting NF-κB in AA mice under therapeutic conditions prolonged their survival, with >50% of mice fully rescued from lethal BMF, and underscoring the important role for NF-κB in regulating additional proinflammatory pathways that mediate BMF in mice.
Herein, we provide evidence that NF-κB–regulated, aberrant CXCR4 expression may be one means by which CD8+ T cells infiltrate the BM during AA progression in mice. The AA mice used in this study represent an additional tool with which to better understand a rare and complex BMF disorder; however, as with all animal models, care must be taken when extrapolating results to the human disease. Further studies of paired samples of peripheral blood and BM from patients with AA will be needed to confirm the relevance of our findings. Determining the full array of chemokine receptors and adhesion molecules that endow pathogenic CD4+ and CD8+ T cells with the ability to traffic to the BM during AA will undoubtedly add to our understanding of the molecular mechanisms that drive this disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the animal care staff for their excellent support, M. Klingbeil for sonicator use, and A. Fauq for DMHEQ.
This study was supported by the Aplastic Anemia and MDS International Foundation, Inc., the American Heart Association, Inc., a Charles H. Hood Foundation Child Health Research Award, the National Institutes of Health, National Cancer Institute (1P01 CA166009-01A1) (L.M.M.), and the National Science Foundation (MRI BD1-1126366) to the Flow Cytometry Core Facility, University of Massachusetts Amherst.
Authorship
Contribution: C.A.K. and G.G.-P. performed experiments, collected, and analyzed data; L.M.M. designed and supervised experiments with contributions from C.A.K. and G.G.-P; and G.G.-P. wrote the manuscript, with critical input from C.A.K. and L.M.M.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lisa M. Minter, 427K Integrated Sciences Bldg, 661 N. Pleasant St, Department of Veterinary and Animal Sciences, University of Massachusetts Amherst, Amherst, MA 01003; e-mail: lminter@vasci.umass.edu.
References
Author notes
C.A.K. and G.G.-P. contributed equally to this study.