Key Points
CLL exosomes exhibit a disease-relevant microRNA signature.
B-cell receptor signaling enhances exosome secretion in CLL that can be antagonized by ibrutinib.
Abstract
Multiple studies show that chronic lymphocytic leukemia (CLL) cells are heavily dependent on their microenvironment for survival. Communication between CLL cells and the microenvironment is mediated through direct cell contact, soluble factors, and extracellular vesicles. Exosomes are small particles enclosed with lipids, proteins, and small RNAs that can convey biological materials to surrounding cells. Our data herein demonstrate that CLL cells release significant amounts of exosomes in plasma that exhibit abundant CD37, CD9, and CD63 expression. Our work also pinpoints the regulation of B-cell receptor (BCR) signaling in the release of CLL exosomes: BCR activation by α-immunoglobulin (Ig)M induces exosome secretion, whereas BCR inactivation via ibrutinib impedes α-IgM-stimulated exosome release. Moreover, analysis of serial plasma samples collected from CLL patients on an ibrutinib clinical trial revealed that exosome plasma concentration was significantly decreased following ibrutinib therapy. Furthermore, microRNA (miR) profiling of plasma-derived exosomes identified a distinct exosome microRNA signature, including miR-29 family, miR-150, miR-155, and miR-223 that have been associated with CLL disease. Interestingly, expression of exosome miR-150 and miR-155 increases with BCR activation. In all, this study successfully characterized CLL exosomes, demonstrated the control of BCR signaling in the release of CLL exosomes, and uncovered a disease-relevant exosome microRNA profile.
Introduction
Chronic lymphocytic leukemia (CLL) is the most prevalent adult leukemia in the western world and remains incurable with current therapies. Understanding the different contributors to pathogenesis in CLL represents a path by which improved therapeutic options can be proposed. Although the pathogenesis of CLL for many years has been attributed to defective apoptosis of tumor cells, robust death is typically noted when these are removed from the body, suggesting a strong role of nurturing in the tumor microenvironment.1 In vivo, CLL cells reside in close contact with T lymphocytes, stromal cells, monocyte-derived nurse-like cells, follicular dendritic cells, and macrophages, collectively referred to as the “microenvironment.” Interactions between these components result in CLL cell trafficking, survival, proliferation, and the increase of the apoptotic threshold, which may be partly dependent on direct physical cell-to-cell contact or mediated through soluble factors. This crosstalk between CLL and the microenvironment is bidirectional; thus, CLL cells are not only being supported by the microenvironment but also are capable of activating and signaling through the secretion of mediators that sustain and promote their survival advantage. In vitro models and gene expression profiles have identified important pathways for the crosstalk between CLL cells and their microenvironment, particularly B-cell receptor (BCR) signaling, which can also contribute to CLL cell survival, proliferation, adhesion, migration, and drug resistance.2-4 CLL can be subdivided into cases with or without somatic mutations in the variable regions of the immunoglobulin heavy chain (IGHV) genes; expression of a mutated IGHV gene generally defines an indolent course, whereas expression of an unmutated IGHV gene is associated with progressive disease.5,6 Unmutated CLL has higher levels of the protein tyrosine kinase ζ-associated protein 70 (ZAP70) and CD38 expression than mutated CLL and can activate key signal transduction pathways, such as spleen tyrosine kinase, Lyn, and Bruton tyrosine kinase (BTK), in response to BCR activation.7-9 Underscoring the relevance of the BCR pathway in disease development and progression, genetic or therapeutic inhibition of these key kinases in this pathway has been shown to induce cytotoxicity in CLL and achieve clinical efficacy.10-12
Communication between cancer cells, including CLL, and their surrounding microenvironment has been well established to preserve tumor survival.13,14 In addition, the release and exchange of secreted extracellular vesicles (EVs) is an alternative means for intercellular communications between tumor and immune cells.15,16 These EVs, including microvesicles and exosomes, have distinct biochemical properties and contain cellular components, such as proteins, peptides, lipids, mRNAs, and microRNAs (miR).17,18 Microvesicles are 100 to 1000 nm in diameter and are generated by shedding from the plasma membrane. Ghosh et al19,20 showed that CLL plasma-derived microvesicles can induce the mammalian target of rapamycin (mTOR)/protein kinase B (AKT)/hypoxia-inducible factor 1-α (HIF-1α) axis and activate AKT through delivery of phospho-receptor tyrosine kinase Axl to CLL bone marrow stromal cells. Moreover, microRNAs previously associated with CLL are found circulating in CLL patient plasma and in plasma-derived microvesicles.21,22 In contrast to microvesicles, exosomes are smaller (50-100 nm) and are considered to initiate from the late endosomal membrane, which gives rise to intracellular multivesicular bodies that later fuse with the plasma membrane and release the exosomes to the extracellular environment.23,24 Differential isolation of microvesicles vs exosomes is performed based on distinct ultracentrifugation techniques.25,26 Exosomes can be found in several bodily fluids, including plasma, serum, and urine, and from several cell types and tissues.26 The secretion of exosomes has been reported to be regulated by increased intracellular calcium levels25 or activation of cell surface receptors27 or stress responses,28 implying that different compositions of exosomes may be produced according to the state of the cell. Emerging evidence also suggests that tumor cells release large amount of exosomes relative to normal cells and that the amount of exosomes released increases as the disease advances.19,29 Moreover, exosome functions in recipient cells are associated with tumor immune response,30 metastasis,31 tumorigenesis,29 angiogenesis, drug resistance,32 and changes in signaling cascades, such as the mTOR/AKT,19,33 extracellular signal-regulated kinase 1/2,34 Wnt,35,36 and transforming growth factor-β/Smad pathways.37 Given the absence of studies of plasma exosomes in CLL and the increasing evidence regarding the importance of these in cancer, we sought to determine whether plasma-derived exosomes from CLL patients are unique relative to microRNA cargo and if the release of these exosomes are influenced by signaling pathways important for CLL survival.
Our study herein identifies unique properties of CLL exosomes including a distinct exosome microRNA signature and prominent CD37 expression on the exosome surface. Further analyses demonstrate that CLL cells secrete more exosomes in plasma than B cells from healthy donors and that the amount of exosomes released in plasma is not correlated with absolute lymphocyte counts but rather regulated by the BCR signaling pathway.
Materials and methods
Patient sample processing, cell culture, chemical reagents, and cell treatment
For all studies, written informed consent was obtained in accordance with the Declaration of Helsinki to procure cells from patients diagnosed with CLL as defined by the modified International Workshop on CLL (iwCLL) 2008 criteria.38 Approval was obtained from The Ohio State University (OSU) institutional review board. CD19+ B cells from CLL patients and healthy donors were isolated and maintained in culture as previously described.39 Cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) and l-glutamine and antibiotics (Life Technologies, Grand Island, NY). For in vitro exosome production experiments, purified CLL cells were cultured in AIM V Serum Free Medium (https://www.lifetechnologies.com/us/en/home/life-science/cell-culture/mammalian-cell-culture/specialty-media/t-cell-media/aim-v-medium.html) to prevent exogenous exosome contamination from FBS. AIM V medium has been tested to sustain the growth of a variety of cell types without supplement of FBS. α-Immunoglobulin (Ig)M stimulation of CLL cells was performed with 10 µg/mL IgM-coated plates with overnight incubation at 37°C. Goat α-human IgM (Fc5μ) was purchased from Jackson ImmunoResearch (West Grove, PA). Ibrutinib (PCI-32765) and idelalisib were obtained commercially (Selleck Chemicals, Houston, TX), as was fludarabine (Sigma-Aldrich, St. Louis, MO). CLL cells were treated with 1 μM ibrutinib for 1 hour and then washed twice with AIM V medium to prevent nonselective kinase inhibition.
Exosome isolation and nanoparticle tracking analysis
Exosomes were isolated by differential ultracentrifugation according to the published procedure26 (see supplemental Methods for details, available on the Blood Web site). The resulting exosome pellet was subjected to size and concentration measurement by NanoSight NS300 (Malvern Instruments, Westborough, MA) at the OSU Comprehensive Cancer Center (OSUCCC) Analytical Cytometry Shared Resource. NanoSight with nanoparticle tracking analysis (NTA) software visualizes and analyzes particles in liquids from 10 to 2000 nm by relating the rate of Brownian motion to particle size.40 The light scattered by the particles with laser illumination is captured by a digital camera, and the motion of each particle is tracked from frame to frame. The rate of particle movement is related to a sphere-equivalent hydrodynamic radius as calculated through the Stokes-Einstein equation. The measurement of exosome concentration performed in this study calculates particle size on a particle-by-particle basis in 5 30-second videos to provide accuracy and statistics for each analysis.
Immunoblotting
Exosomes were lysed and sonicated, and protein concentration was quantified by the detergent compatible (DC) protein assay from BioRad (http://www.bio-rad.com/en-us/product/dc-protein-assay) as previously described.41 Twenty-five micrograms lysate was resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to the polyvinylidene fluoride (PVDF) membrane, and blotted with specific primary and secondary antibodies (Sigma-Aldrich) accordingly. Primary antibodies used for immunoblotting for CD63 (H-193) and CD9 (C-4) were purchased from Santa Cruz Biotechnology (Dallas, TX).
Analysis of exosomes bound to beads by flow cytometry
Although the measurement of larger microvesicles has been reported to be feasible by flow cytometry, such methodology is not adaptable to smaller exosomes. We therefore adopted a technique to visualize purified exosomes bound to beads of a size that can be detected on a cytometer (see supplemental Methods for details).26 The exosome-bead complex was then stained with fluorophore-coupled primary antibodies or isotype controls and analyzed on a Gallios flow cytometer (Beckman Coulter, Brea, CA). Voltage was adjusted to detect beads from the dot plot of forward and side scatter and set to a linear scale. From this plot, single beads and clumps of 2 and 3 beads could be observed. Only single beads were gated for fluorescence signal analysis. Fluorescence obtained from the specific antibody and isotype controls was compared and analyzed by Kaluza software (Beckman Coulter).
microRNA isolation, quantitative real-time PCR, small RNA analysis, and exosome microRNA profiling
CD19+ purified B cells or exosomes were homogenized with TRIzol reagent (Life Technologies) or QIAzol lysis reagent (Qiagen, Valencia, CA) and isolated for microRNA by miRNeasy Mini or Micro Kit (Qiagen). cDNA was prepared with microRNA-specific primers and TaqMan MicroRNA Reverse Transcription Kit (Life Technologies). microRNA expression was measured by quantitative real-time polymerase chain reaction (PCR) with TaqMan microRNA assays for miR-150, miR-155, miR-223, miR-29a-c, RNU44, and miR-192 (Life Technologies) and detected by the ViiA 7 Real-Time PCR System (Life Technologies). RNU44 and miR-192 are used as housekeeping microRNAs for normalization of cellular and exosome microRNAs, respectively (supplemental Figure 1). Isolated small RNA from CLL exosomes was submitted to the OSUCCC Genomic Shared Resource for the analysis and quantitation of small RNA samples of 6 and 150 nt in size. Small RNA samples were size fractionated, purified, and concentrated by using a gel-based fractionation system, Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA). Exosome microRNA collected from 1 mL plasma from 69 CLL patients and 15 healthy donors was also profiled by the nCounter miRNA Expression Assay (NanoString Technologies, Seattle, WA) at the Genomic Shared Resource. The nCounter Analysis System uses color-coded molecular barcodes to hybridize directly to a target molecule in solution and single-molecule imaging to detect and count hundreds of unique transcripts in a single reaction for data collection. Unlike other methods, this protocol does not include any amplification steps that might introduce bias to the results. Results were analyzed to detect differentially expressed microRNAs between CLL and healthy donors. We optimized our methods to use as little as 1 mL of plasma, achieving RNA quality and microRNA profile results equivalent to those obtained from 10 mL of plasma (supplemental Figure 2).
Statistical analysis
nCounter microRNA expression assay count data were normalized in 2 steps. First, technical normalization was performed according to guidelines provided by NanoString Technologies. Positive controls were used to adjust the microRNA counts for technical assay variation. Then, biological normalization was performed to correct for differences in sample abundances. The geometric mean of the most stable 20 microRNAs was used to normalize the data. A list of the 20 most stable exosome microRNAs is presented in supplemental Table 1. This approach, which was originally developed to provide accurate and data-driven normalization factors to quantitative real-time PCR data,42 was then applied to RNA sequencing count data to identify the most variable genomic features43 and successfully usedto normalize the nCounter miRNA assay count data as well.44,45
Differences in exosome concentration between healthy donors and CLL patients, as well as IGHV-mutated and -unmutated CLL patients, were examined using 2-sample t tests with unequal variances. Paired t tests or mixed effects models were used to assess differences in exosome concentration/production between conditions within patient samples (eg,, ibrutinib in the presence of α-IgM stimulation vs stimulation alone). Data were log-transformed to improve normality. The Pearson correlation coefficient between exosome concentrations in CLL plasma with absolute lymphocyte counts was calculated, with the 95% confidence interval (CI). All exosome comparisons were performed using SAS/STAT software, version 9.2 (SAS Institute, Cary, NC).
Results
Characterization of plasma-derived exosomes in CLL
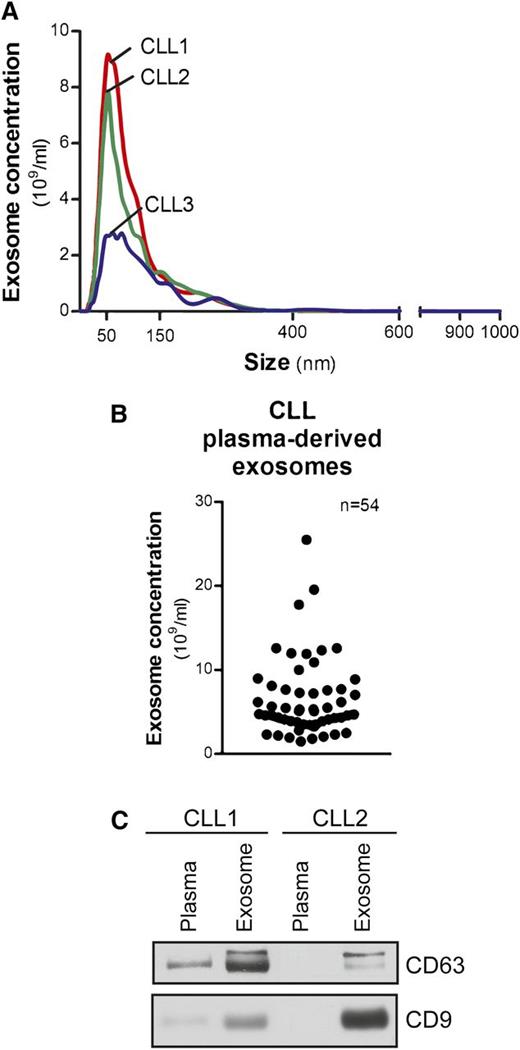
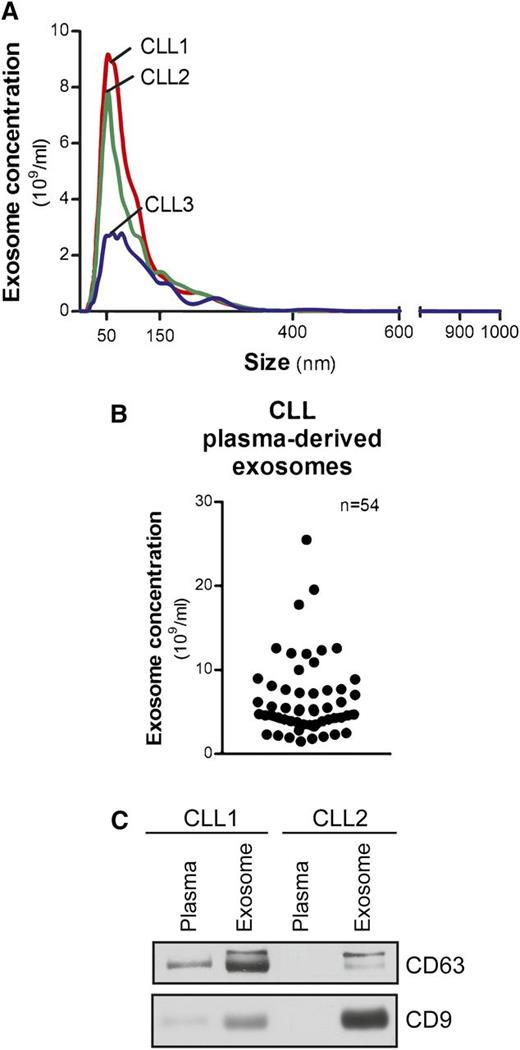
Exosomes from CLL patient plasma, isolated by differential centrifugation, were measured within 50 to 150 nm in diameter (Figure 1A). Concentrations ranged from 1.5 to 25.5 × 109/mL and were not associated with patients’ absolute lymphocyte counts (Figure 1B). Exosome markers, including the tetraspanins CD63 and CD9,23 were evaluated by immunoblot to demonstrate the purity and quality of our isolation technique (Figure 1C).
Characterization of CLL exosomes in plasma. (A) Exosomes were purified by differential centrifugation from CLL patient plasma and subjected to NTA measurement for concentration and size distribution. The representative plot of each CLL sample present here was generated from the average of 5 30-second videos. (B) Dot plot showing the range of exosome concentrations detected in CLL plasma (n = 54). (C) Isolated CLL plasma-derived exosomes were immunoblotted for exosome markers CD63 and CD9. Purified exosomes showed enriched CD63 and CD9 expression compared with plasma samples.
Characterization of CLL exosomes in plasma. (A) Exosomes were purified by differential centrifugation from CLL patient plasma and subjected to NTA measurement for concentration and size distribution. The representative plot of each CLL sample present here was generated from the average of 5 30-second videos. (B) Dot plot showing the range of exosome concentrations detected in CLL plasma (n = 54). (C) Isolated CLL plasma-derived exosomes were immunoblotted for exosome markers CD63 and CD9. Purified exosomes showed enriched CD63 and CD9 expression compared with plasma samples.
Surface antigens CD63, CD9, and CD37 are abundant on CLL exosomes
Many cell types are reported to release exosomes, in particular cells of the hematopoietic lineage including platelets,25 T cells,46 B cells,47 natural killer cells,48 and dendritic cells.49 Therefore, we sought to determine the origin of exosomes in CLL plasma by assessing expression of cell surface antigens using flow cytometry. Exosomes are too small to be analyzed by direct flow cytometry, so for our experiments, purified exosomes were bound to beads of a size that is in the detection range of a flow cytometer. The exosomes were then stained with fluorophore-conjugated primary antibodies and analyzed by a flow cytometer. The results obtained from flow cytometry were consistent with our immunoblots while CLL plasma-derived exosomes were detected with the enriched exosome proteins such as CD63 and CD9 (Figure 2A). Moreover, CLL plasma-derived exosomes expressed predominantly CD37, CD63, and CD9 but not CD3, CD56, or CD41, suggesting that these exosomes are primarily released from B-CLL cells but not T cells or natural killer cells (Figure 2B). Activated platelets have been shown to produce high levels of EVs, and CD41, a platelet marker, has been detected mainly on microvesicles but not exosomes.25 Consistent with previous research, our data show that CLL plasma-derived exosomes contain minimal CD41, indicating that CLL plasma contains negligible amounts of platelet-derived exosomes.
Determination of surface protein expression on CLL exosomes by flow cytometry. (A) Purified CLL exosomes from plasma were first bound to beads with a size that can be detected by direct sorting. Then the exosome-bead complex was stained with fluorophore-conjugated primary antibodies or matched isotype controls and analyzed by flow cytometry. CLL exosomes expressed abundant levels of CD63, CD9, and CD37 but not CD41, CD3, or CD56. (B) The dot plots present a range of expression of each surface antigen on exosomes isolated from 12 CLL patients.
Determination of surface protein expression on CLL exosomes by flow cytometry. (A) Purified CLL exosomes from plasma were first bound to beads with a size that can be detected by direct sorting. Then the exosome-bead complex was stained with fluorophore-conjugated primary antibodies or matched isotype controls and analyzed by flow cytometry. CLL exosomes expressed abundant levels of CD63, CD9, and CD37 but not CD41, CD3, or CD56. (B) The dot plots present a range of expression of each surface antigen on exosomes isolated from 12 CLL patients.
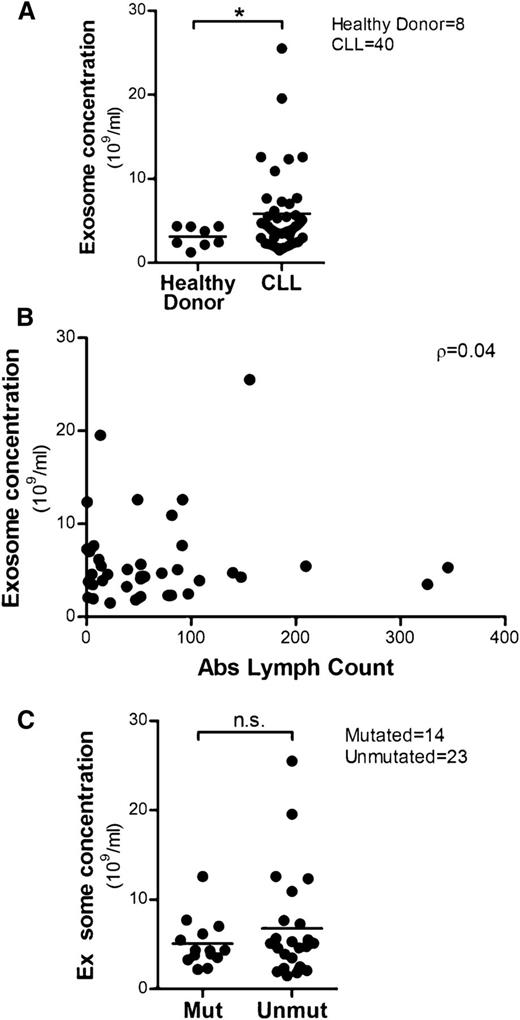
CLL patients release more exosomes in plasma compared with healthy donors
Cancer cells have been shown to release high levels of exosomes that carry tumor-specific antigens or proteins to communicate with the microenvironment to promote disease progression.50,51 Therefore, we isolated plasma-derived exosomes from CLL patients (n = 40) and healthy donors (n = 8) and compared exosome concentrations by NanoSight. There was a significantly higher level of exosomes in CLL plasma compared with healthy donors (*P = .026; Figure 3A). We then determined that exosome concentrations in CLL patient plasma were not significantly correlated with absolute lymphocyte counts (ρ = 0.04; 95% CI: −0.28, 0.35; P = .800), suggesting the release of exosomes is regulated by yet-to-be determined mechanisms in CLL (Figure 3B). It has also been shown that the secretion of exosomes can be induced by different extracellular stimuli25,52 or activation of receptors27,53 in many cell types. In CLL, unmutated IGHV is associated with a poor clinical course5,6 and strong activation of BCR signaling transduction, indicating that BCR signaling plays an important role in the proliferation and maintenance of the malignant B cells.9,54 Thus, we compared plasma exosome concentrations in IGHV-unmutated and -mutated CLL to examine the association between mutational status and exosome secretion. Although overall there was no statistically significant difference in exosome concentration between unmutated and mutated CLL, ∼52% (12 of 23) of unmutated patients exhibited an exosome concentration >5.0 × 109/mL compared with only 36% (5 of 14) of mutated patients (Figure 3C).
High levels of exosomes are detected in CLL plasma and their concentrations are not correlated with absolute lymphocyte counts. (A) Exosomes were isolated from 40 CLL patients’ plasma and 8 healthy donors’ plasma, and the exosome concentrations were determined by NTA. Plasma from CLL patients showed higher concentrations of exosomes compared with plasma from healthy donors (*P = .026). (B) No significant correlation between plasma exosome concentrations and absolute lymphocyte counts was determined, as calculated by Spearman’s correlation coefficient (n = 39, ρ = 0.04). (C) Plasma exosome concentrations in IGHV-unmutated CLL was not significantly higher than in IGHV-mutated CLL; however, there is a trend in which more unmutated CLL patients exhibit elevated exosome concentrations vs IGHV-mutated CLL patients.
High levels of exosomes are detected in CLL plasma and their concentrations are not correlated with absolute lymphocyte counts. (A) Exosomes were isolated from 40 CLL patients’ plasma and 8 healthy donors’ plasma, and the exosome concentrations were determined by NTA. Plasma from CLL patients showed higher concentrations of exosomes compared with plasma from healthy donors (*P = .026). (B) No significant correlation between plasma exosome concentrations and absolute lymphocyte counts was determined, as calculated by Spearman’s correlation coefficient (n = 39, ρ = 0.04). (C) Plasma exosome concentrations in IGHV-unmutated CLL was not significantly higher than in IGHV-mutated CLL; however, there is a trend in which more unmutated CLL patients exhibit elevated exosome concentrations vs IGHV-mutated CLL patients.
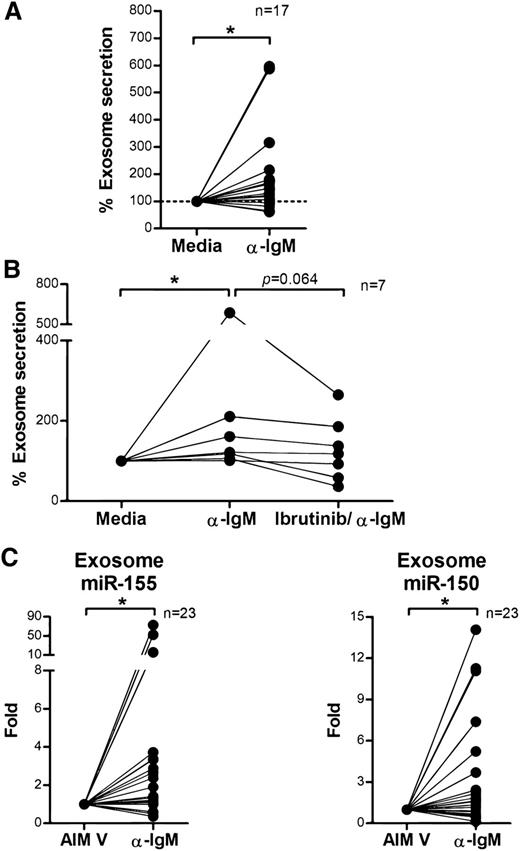
BCR signaling enhances the secretion of exosomes in CLL
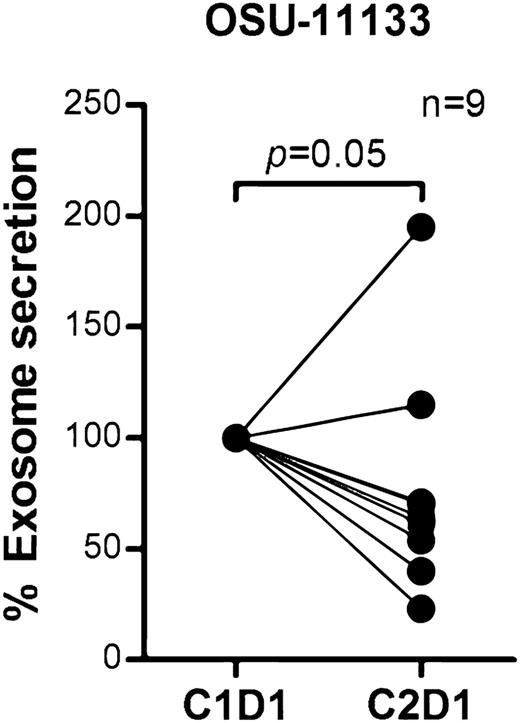
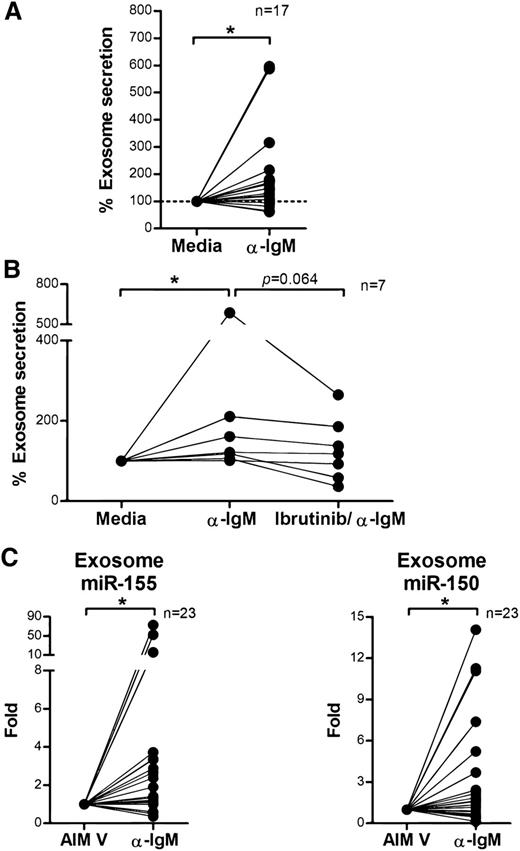
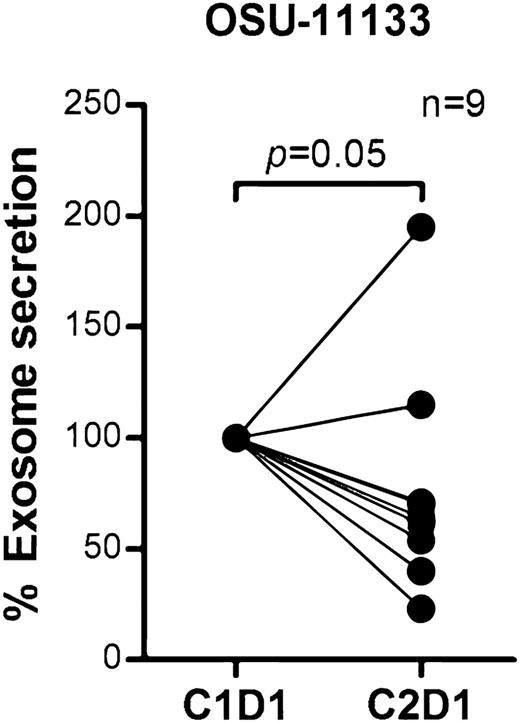
BCR signaling plays an important role in CLL pathogenesis and cell survival and therefore we sought to investigate whether BCR signaling controls exosome secretion in CLL. We isolated CD19+ CLL cells (n = 17), cultured them in AIM V medium with or without stimulation via plate-bound α-IgM for 24 hours, and measured exosome concentrations in the supernatants. Stimulation with α-IgM significantly increased exosome release from CLL cells (estimated fold change = 1.56; 95% CI: 1.11, 2.18; P = .013; Figure 4A). Next, we validated this effect by therapeutic inhibition of BCR signaling with ibrutinib. Ibrutinib is a covalently bound BTK inhibitor that specifically and effectively prevents BCR signaling transduction in vitro and in vivo.55,56 We treated purified CD19+ CLL cells with ibrutinib for 1 hour, washed away the drug to prevent any off-target effect, and then incubated the cells with or without α-IgM for 24 hours and measured exosomes in supernatants as above. Among the 7 of 12 CLL patient samples that responded to α-IgM stimulation, BCR inactivation via ibrutinib decreased the exosome release 3% to 66% (P = .064; Figure 4B). Next, we purified exosomes from plasma samples collected on an ibrutinib phase 2 clinical trial (OSU-11133; #NCT01589302) and compared exosome concentrations before (cycle 1, day 1) vs after (cycle 2, day 1) ibrutinib treatment. Our data show that the concentration of exosomes in plasma was significantly decreased in CLL patients after 28 days of ibrutinib therapy (P = .05; Figure 5). Moreover, we also examined whether other CLL therapies, including the phosphoinositide 3-kinase inhibitor, idelalisib, and chemotherapy, fludarabine, contribute to CLL exosome release. Our results showed that idelalisib but not fludarabine significantly downregulated exosome secretion in vitro, underlining that the BCR-phosphoinositide 3-kinase pathway is in control of exosome release in CLL (supplemental Figure 3).
BCR signaling regulates exosome secretion and exosome microRNA expression in CLL. (A) CD19+ CLL cells were purified and cultured in AIM V medium with or without α-IgM stimulation for 24 hours. CLL-derived exosomes were isolated from cell culture supernatant, and the exosome concentration was measured by NTA. α-IgM stimulation significantly increased exosome secretion from CLL cells (*P = .013). (B) CD19+ CLL cells were isolated and first treated with or without 1 μM ibrutinib for 1 hour and then washed to remove the drug. Cells were then stimulated with α-IgM for 24 hours, and exosomes were isolated from the supernatant for measurement of exosome concentration by NTA. In the 7 of 12 CLL cells that responded to α-IgM stimulation (*P = .011), ibrutinib treatment decreased α-IgM-induced exosome release (P = .064). (C) Exosome microRNA was also isolated from the supernatant of cells incubated with or without α-IgM, and analyzed for miR-150 and miR-155 expression. Both miR-150 and miR-155 showed significantly higher levels of expression after α-IgM stimulation (*P = .042 and *P = .014, respectively).
BCR signaling regulates exosome secretion and exosome microRNA expression in CLL. (A) CD19+ CLL cells were purified and cultured in AIM V medium with or without α-IgM stimulation for 24 hours. CLL-derived exosomes were isolated from cell culture supernatant, and the exosome concentration was measured by NTA. α-IgM stimulation significantly increased exosome secretion from CLL cells (*P = .013). (B) CD19+ CLL cells were isolated and first treated with or without 1 μM ibrutinib for 1 hour and then washed to remove the drug. Cells were then stimulated with α-IgM for 24 hours, and exosomes were isolated from the supernatant for measurement of exosome concentration by NTA. In the 7 of 12 CLL cells that responded to α-IgM stimulation (*P = .011), ibrutinib treatment decreased α-IgM-induced exosome release (P = .064). (C) Exosome microRNA was also isolated from the supernatant of cells incubated with or without α-IgM, and analyzed for miR-150 and miR-155 expression. Both miR-150 and miR-155 showed significantly higher levels of expression after α-IgM stimulation (*P = .042 and *P = .014, respectively).
Exosome concentrations were decreased in CLL patients after receiving ibrutinib therapy. Exosomes were isolated from plasma collected before (cycle 1, day 1) and after ibrutinib treatment (cycle 2, day 1) in a phase 2 clinical trial (OSU-11133; #NCT01589302). Plasma exosome concentration in CLL patients after receiving ibrutinib therapy was significantly reduced compared with pretreatment (P = .05).
Exosome concentrations were decreased in CLL patients after receiving ibrutinib therapy. Exosomes were isolated from plasma collected before (cycle 1, day 1) and after ibrutinib treatment (cycle 2, day 1) in a phase 2 clinical trial (OSU-11133; #NCT01589302). Plasma exosome concentration in CLL patients after receiving ibrutinib therapy was significantly reduced compared with pretreatment (P = .05).
microRNA profiling of exosomes reveals a distinct CLL signature
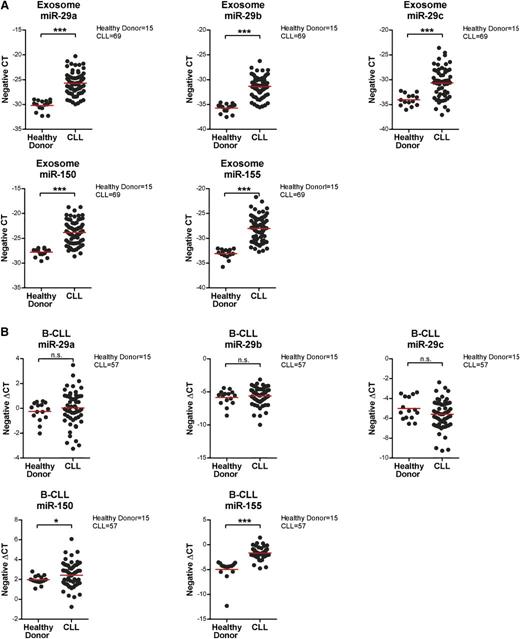
Communication between cells and the microenvironment has been shown by multiple means, including exosome-mediated transfer of protein, microRNA, or mRNA to the recipient cells.34,51,57,58 microRNAs are small noncoding RNA molecules (∼20 nucleotides) that affect protein expression either by posttranscriptional silencing via transcript degradation and/or inhibition of translation. It has also been demonstrated that many microRNAs are highly associated with CLL disease progression and prognosis,59,60 and recent studies have suggested that circulating microRNAs in CLL plasma can be potential prognostic biomarkers.21,22 Thus, we first examined the microRNA content in CLL plasma-derived exosomes and then profiled the CLL exosome microRNAs, exploiting small RNA analysis to distinguish the microRNAs from other small RNA species in CLL exosomes. Our results indicate that CLL exosomes have a rich content of microRNA and a small portion of transfer RNAs and small ribosomal RNAs (supplemental Figure 4). Next, we conducted microRNA profiling via the nCounter miRNA expression assay on plasma-derived exosomes from 69 CLL patients and 15 healthy donors. These results identified a distinct microRNA signature in CLL plasma-derived exosomes, including upregulation of miR-150, miR-155, and miR-29 family members and downregulation of miR-223 (Table 1). Cellular expression of these microRNAs has previously been associated with CLL disease.59,61-64 Moreover, other differentially expressed microRNAs identified in the CLL exosome microRNA profile have been shown to be abnormally expressed and implicated in the development of other types of cancer.65-69 We then confirmed the results of exosome microRNA profiling by quantitative real-time PCR and verified that levels of miR-150, miR-155, and miR-29a-c are significantly higher in circulating CLL exosomes compared with exosomes from healthy donors (P < .001 for all comparisons; Figure 6A). Interestingly, when examining microRNA expression levels in the corresponding CLL and normal B cells, miR-150 and miR-155 were also significantly higher in CLL vs normal, but there were no differences in expression levels of miR-29 family members (P = .029 and P < .001, respectively; Figure 6B). This observation suggests the presence of active regulatory pathways that affect exosome content between CLL and normal B cells.
Confirmation of exosome and cellular expression of identified microRNAs by quantitative real-time PCR. (A) Exosomes were isolated from 1 mL frozen plasma samples from 69 CLL patients and 15 healthy donors. Isolated exosome microRNA was examined by quantitative real-time PCR to determine the expression of miR29a-c, miR-150, and miR-155. All these microRNAs were expressed at significantly higher levels in CLL patients compared with healthy donors (***P < .001 for all comparisons). The expression data shown here are in a scale of negative CT (cycles required to reach threshold of detection), where a higher number represents higher expression and vice versa. (B) microRNAs were also isolated from the corresponding CD19+ CLL or normal B cells and analyzed by quantitative real-time PCR. Cellular levels of miR-150 and miR-155 were significantly higher in tumor cells from CLL patients vs normal B cells from healthy donors (*P = .029 and ***P < .001, respectively). There were no significant differences in the expression of cellular miR-29a-c between CLL patients and healthy donors.
Confirmation of exosome and cellular expression of identified microRNAs by quantitative real-time PCR. (A) Exosomes were isolated from 1 mL frozen plasma samples from 69 CLL patients and 15 healthy donors. Isolated exosome microRNA was examined by quantitative real-time PCR to determine the expression of miR29a-c, miR-150, and miR-155. All these microRNAs were expressed at significantly higher levels in CLL patients compared with healthy donors (***P < .001 for all comparisons). The expression data shown here are in a scale of negative CT (cycles required to reach threshold of detection), where a higher number represents higher expression and vice versa. (B) microRNAs were also isolated from the corresponding CD19+ CLL or normal B cells and analyzed by quantitative real-time PCR. Cellular levels of miR-150 and miR-155 were significantly higher in tumor cells from CLL patients vs normal B cells from healthy donors (*P = .029 and ***P < .001, respectively). There were no significant differences in the expression of cellular miR-29a-c between CLL patients and healthy donors.
Levels of exosome miR-150 and miR-155 increased on BCR activation
Overexpression of miR-155 in CLL is associated with more aggressive disease and poor response to chemoimmunotherapy.21,59 It also has been shown that BCR activation via α-IgM stimulation can induce cellular miR-155 expression in Ramos cells70 and that transfection of miR-155 can enhance the sensitivity of CLL cells to BCR ligation.61 These observations suggest a strong association between miR-155 and BCR activation. Given our results demonstrating that α-IgM stimulation increases exosome secretion from CLL cells, we examined expression levels of exosome microRNA following α-IgM treatment using quantitative real-time PCR. We found no statistically significant differences in exosome miR-29a-c expression following α-IgM stimulation (data not shown). However, expression of exosome miR-150 and miR-155 was significantly increased following α-IgM treatment (P = .042 and P = .014, respectively), in concert with our results of higher expression of exosome miR-150 and miR-155 in CLL (Figure 4C).
Discussion
This study for the first time characterizes the presence of distinct plasma-derived exosomes in CLL with abundant surface expression of CD9, CD63, and CD37 and modest expression of CD41, CD56, and CD3 via flow analysis and immunoblotting. Our results show that CLL cells release significantly higher levels of exosomes in plasma compared with B cells from healthy donors and that plasma exosome concentration in CLL is not associated with the absolute lymphocyte count. These findings suggest that CLL exosomes are predominantly released from B lymphocytes and the release of exosomes is regulated rather than spontaneously secreted. We demonstrate that CLL plasma-derived exosomes have unique microRNA expression compared with normal controls. In addition, BCR activation in CLL by α-IgM stimulation induces the secretion of exosomes, whereas inhibition of BCR signaling via the BTK inhibitor ibrutinib abrogated α-IgM-induced CLL exosome release. The association of exosome release with BCR signaling was also indicated in vivo using serial plasma samples from relapsed CLL patients receiving ibrutinib therapy. Moreover, expression of miR-150 and miR-155 in exosomes was significantly elevated in CLL-derived exosomes vs normal B cells and further increased with α-IgM stimulation. These data implicate the BCR signaling pathway in the control of CLL exosome secretion and its conveyance of the disease-relevant microRNAs miR-150 and miR-155. Collectively, these findings provide impetus for further mechanistic investigation of exosomes in pathogenic signaling processes in CLL and the surrounding microenvironment.
Although our findings are unique and represent the first description of plasma-released exosomes in CLL, they are complementary to previous work examining the plasma content of microRNA. For example, Ferrajoli and colleagues21 demonstrated that plasma miR-155 correlated with response to both chemoimmunotherapy with fludarabine, cyclophosphamide, and rituximab and also immune therapy with lenalidomide. Additionally, preliminary studies within this manuscript identified that larger microvesicles isolated in a different manner than exosomes also contained miR-155. Work by others has shown that the microvesicle-derived miR-155 interaction with TLR7 signaling is physiologically relevant as well.71 The complex extracellular communication of key microRNA, proteins, and other RNA products is just being appreciated, and future studies elucidating further the differential regulation of these are warranted.
Moving forward, it would be intriguing to examine whether other extracellular stimuli that have been shown to produce prosurvival signals in CLL including CD40L, interleukin-4, or B-cell activating factor (BAFF) also contribute to the secretion of CLL exosomes and the inclusion of selective microRNAs. Additionally, examination of the contribution of CLL exosome number and content in predicting response to different therapeutic agents would be of interest. In this regard, the methodology established herein combined with new next-generation sequencing techniques will provide the ability to quantify microRNAs in 1 mL of plasma or even less. Investigations by our group examining the impact of plasma-derived exosomes and microRNA content on response to targeted therapies are ongoing. In addition to the prognosis, future work will also examine the potential resistant mechanisms to therapies contributed by transfer of CLL exosomes.
In conclusion, we optimized the methodology and techniques to successfully characterize CLL exosomes with detection of surface protein expression via flow cytometry, assessment of exosome proteins via immunoblotting, quantification of exosomes via NanoSight, and identification of the microRNA profile via the nCounter microRNA expression assay. This endeavor provides a new understanding of CLL exosomes and builds the foundation for future studies. We also demonstrated for the first time the role of BCR signaling in controlling the release of exosomes, as well as the upregulation of exosome miR-150 and miR-155 in CLL. Moreover, the identified CLL exosome microRNA signature can be explored further to uncover exosome function and its relevance to CLL disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a Leukemia & Lymphoma Society SCOR grant, the D. Warren Brown Foundation, and the National Institutes of Health National Cancer Institute grants P50 CA140158, P01 CA81534, and U01 CA076576.
Authorship
Contribution: Y.-Y.Y. planned the research, performed experiments, analyzed data, and drafted the paper; A.M.L., L.Y., and H.G.O. were involved in the bioinformatics and statistical analysis and reviewed drafts; K.M. and J.C.B. contributed patients and collected data; and J.C.B. and A.J.J. planned and supervised the research, analyzed the data, reviewed drafts, and obtained funding for the research work.
Conflict-of-interest disclosure: J.C.B. served as an unpaid consultant for Pharmacyclics, Inc. The remaining authors declare no competing financial interests.
Correspondence: John C. Byrd, 455B OSUCCC, 410 West 12th Ave, The Ohio State University, Columbus, OH 43210; e-mail: john.byrd@osumc.edu; or Amy J. Johnson, 455C OSUCCC, 410 West 12th Ave, The Ohio State University, Columbus, OH 43210; e-mail: amy.johnson@osumc.edu.
References
Author notes
A.J.J. and J.C.B. contributed equally to this work.