Key Points
Mouse BM-derived mast cells can dedifferentiate into immature myeloid-like cells after the deletion of the GATA2 DNA binding domain.
Increased expression of C/EBPα is critical for the dedifferentiation of GATA2-deficient mast cells.
Abstract
GATA2 plays a crucial role for the mast cell fate decision. We herein demonstrate that GATA2 is also required for the maintenance of the cellular identity in committed mast cells derived from mouse bone marrow (BMMCs). The deletion of the GATA2 DNA binding domain (GATA2ΔCF) in BMMCs resulted in a loss of the mast cell phenotype and an increase in the number of CD11b- and/or Ly6G/C-positive cells. These cells showed the ability to differentiate into macrophage- and neutrophil-like cells but not into eosinophils. Although the mRNA levels of basophil-specific genes were elevated, CD49b, a representative basophil marker, never appeared on these cells. GATA2 ablation led to a significant upregulation of C/EBPα, and forced expression of C/EBPα in wild-type BMMCs phenocopied the GATA2ΔCF cells. Interestingly, simultaneous deletion of the Gata2 and Cebpa genes in BMMCs restored the aberrant increases of CD11b and Ly6G/C while retaining the reduced c-Kit expression. Chromatin immunoprecipitation assays indicated that GATA2 directly binds to the +37-kb region of the Cebpa gene and thereby inhibits the RUNX1 and PU.1 binding to the neighboring region. Upregulation of C/EBPα following the loss of GATA2 was not observed in cultured mast cells derived from peritoneal fluid, whereas the repression of c-Kit and other mast cell-specific genes were observed in these cells. Collectively, these results indicate that GATA2 maintains cellular identity by preventing Cebpa gene activation in a subpopulation of mast cells, whereas it plays a fundamental role as a positive regulator of mast cell-specific genes throughout development of this cell lineage.
Introduction
A zinc finger transcription factor, GATA2, has been shown to be essential for early mast cell development. A previous study showed that GATA2-deficient yolk sac cells failed to differentiated into mast cells under the interleukin 3 (IL-3) and stem cell factor (SCF)-supplemented culture conditions.1 The GATA2 expression is also detected in the differentiated mast cells in peripheral tissues.2 A recent study by our group showed that the GATA2 mRNA level was significantly increased during the differentiation of bone marrow-derived mast cells (BMMCs) in mice, suggesting a pivotal role for this factor in differentiated mast cells.3
GATA1, another GATA transcription factor family member, is coexpressed with GATA2 in mast cells. Recently, we reported that conditional ablation of GATA1 in adult mice exhibited a minimal effect on the cell number and distribution of peripheral tissue mast cells.4 Quantitative chromatin immunoprecipitation (qChIP) assays and silent interfering RNA-mediated knockdown experiments revealed that both GATA1 and GATA2 regulate the expression of a series of known GATA factor downstream genes in BMMCs.5-7 Interestingly, however, inducible deletion of GATA2 DNA-binding domain C-finger (CF) in BMMCs resulted in a significant reduction in the number of the c-Kit/FcεRIα double-positive (DP) cells, whereas the deletion of GATA1 did not significantly affect the frequency of DP cells or the cell morphology or viability.4 These results suggest that GATA2 plays a more important role than GATA1 for maintaining the mast cell identity in BMMCs. In addition to GATA1 and GATA2, several other transcription factors including microphthalmia-associated transcription factor (MITF), runt related transcription factor 1 (RUNX1), T cell acute lymphocytic leukemia 1, and spleen focus forming virus (SFFV) proviral integration oncogene (PU.1) have been demonstrated to be involved in mast cell development.8-11 At present, it is virtually unknown how the interplay of these transcription factors affects the individual factors' functions during mast cell differentiation. In addition, it is not known which factor has the most significant effect on mast cell differentiation.
The present study aimed to define the precise role of GATA2 in mast cell differentiation by conditionally deleting GATA2 CF (hereafter referred to as ΔCF) in BMMCs. We found that GATA2-deficient BMMCs rapidly dedifferentiated into the CD11b+ cells and acquired a potential to differentiate into macrophage- and neutrophil-like cells, but not into eosinophils, in the presence of myeloid cytokines. The GATA2 CF deletion resulted in an immediate and significant increase in the expression of CCAAT/enhancer binding protein α (C/EBPα), a key transcription factor required for myeloid lineage determination.12-14 Finally, we show that C/EBPα expression was rarely upregulated by the loss of GATA2 in cultured mast cells derived from peritoneal fluid (PMCs), while decreased expression of c-Kit and other mast cell-specific genes was observed in the GATA2-deficient PMCs. These results indicate that GATA2 maintains cellular identity by preventing Cebpa gene activation in a subpopulation of mast cells, whereas it plays a fundamental role as a positive regulator of mast cell-specific genes throughout development of this cell lineage.
Methods
Mice
Gata2flox/flox mice15 were kindly provided by S. A. Camper (University of Michigan). Cebpaflox/flox mice16 were purchased from Jackson Laboratories (stock no. 006447). The 4-hydroxytamoxifen (4-OHT)-inducible Rosa26CreERT2 mice were kindly provided by Anton Berns (Netherlands Cancer Institute). The Gata2 knockout phenotype was examined in BMMCs prepared from homozygous male mice expressing CreERT2 (supplemental Figure 1 available on the Blood Web site). Gata2+/+ mice expressing CreERT2 were used to prepare control BMMCs. The mice were maintained in an animal facility of Takasaki University of Health and Welfare in accordance with institutional guidelines.
RNA extraction and quantitative reverse transcriptase-polymerase chain reaction
Total RNA was extracted from cells using NucleoSpin RNA II (TaKaRa), and 0.5 μg of total RNA was used for the reverse transcription (RT) reactions using a ReverTra Ace qPCR RT kit (Toyobo) according to the manufacturer's instructions. Quantitative reverse transcriptase-polymerase chain reaction (Q-PCR) was performed using Go Taq qPCR master mix (Promega) and an Mx3000P real-time PCR system (Stratagene), as described previously.3,17 The data were normalized to the 18S rRNA expression levels and are shown as the averages ± standard deviation (SD).
Western blotting
Cells were lysed by boiling in 4× sodium dodecyl sulfate (SDS) buffer (312.5 mM Tris-HCl, pH 6.8, 10% SDS [v/v], 25% sucrose [v/v], 0.01% bromophenol blue, and 20% 2-mercaptoethanol). The lysates were resolved by 10% SDS-polyacrylamide gel electrophoresis, and western blot analyses were performed as described previously.3,17
Cytological analyses
Cytospin preparations of the cells (5 × 104 cells) were made using a Shandon Cytospin4 centrifuge (Thermo Electron Corp.) and were stained with Wright-Giemsa stain, toluidine blue, or Diff-Quik (Sysmex). The nucleus/cytoplasm (N/C) ratio was determined by using the Image J software program.
Fluorescence-activated cell sorting analysis
For the fluorescence-activated cell sorting (FACS) analysis, cells were stained with fluorescence-conjugated antibodies for 30 minutes and were analyzed using a FACSCanto II flow cytometer (BD Biosciences) or were sorted using a FACSJazz cell sorter (BD Biosciences).
qChIP assay
The ChIP assay using BMMCs and the quantitative analyses of DNA purified from the ChIP samples were conducted as described previously.17
Statistical analysis
Comparisons between 2 groups were made using the Student t test. The data are presented as means ± SD. For all analyses, statistical significance was defined as a value of P < .05 or P < .01.
Cell culture and next-generation sequencing analysis
The detailed protocols for cell culture, photomicrographs, cell proliferation, cell death and cell cycle assays, transfection of siRNAs, induction of the Cre transgenes in vitro, generation of constructs, production and transduction of retroviruses, phagocytosis assay, and next-generation sequencing analysis are described in the supplemental Methods.
Results
Deletion of GATA2 CF in BMMCs results in a loss of mast cell identity
To investigate the roles of GATA2 in mast cells, BMMCs prepared from Gata2flox/flox::Rosa26CreERT2 mice were treated with 4-OHT on cell culture day 30 (Figure 1A). As reported previously,4 the fraction of DP cells was decreased over time to only 7.9 ± 1.5% of the initial amount on day 9 (Figure 1A). Phase-contrast photomicrographs of the BMMCs on day 10 showed that the ΔCF/ΔCF BMMCs were irregular in size and shape compared with the CreERT2 control BMMCs (Figure 1B). Wright Giemsa and toluidine blue staining showed a paucity of granules in the cytoplasm and an increased N/C ratio in the ΔCF/ΔCF BMMCs (Figure 1B). Although significant changes were observed in the morphology, the GATA2 deletion exerted only a limited effect on the cell number and viability (Figure 1C-D, respectively). However, the FACS analysis with 5-bromo-2′-deoxyuridine and 7-amino-actinomycin D staining showed that there were increases in the subG0/G1, S, and G2/M phase fractions and a substantial decrease in the G0/G1 phase fraction following 4-OHT treatment (Figure 1E). To eliminate the possible contribution of immature progenitor cells contained in the starting cell population, we sorted the DP cell fraction before 4-OHT treatment. As expected, the DP cell population was decreased, similar to the decrease observed in the unsorted BMMCs (Figure 1F). Collectively, these data suggest that the DP BMMCs were dedifferentiated into immature cells by the GATA2 deletion.
The deletion of GATA2 CF results in a loss of mast cell identity in mouse bone marrow-derived mast cells. (A) (Left) Representative FACS data for BMMCs prepared from Gata2flox/flox::Rosa26CreERT2 mice after 0, 3, 6, and 9 days of 4-OHT treatment (D0, D3, D6, and D9, respectively). (Right) Average percentages and SD of the DP cells within the gates examined on days 0, 3, 5, 7, and 9 of the 4-OHT treatment (D0, D3, D5, D7, and D9, respectively). N = 9. (B) Representative bright field (BF) phase contrast photomicrographs (left; original magnification, ×200) and cytospin preparations (center and right; original magnification, ×400) of BMMCs prepared from Gata2+/+::Rosa26CreERT2 (CreERT2) and Gata2flox/flox::Rosa26CreERT2(ΔCF/ΔCF) mice treated with 0.5 μM of 4-OHT for 10 days. For cytospin preparations, the cells were stained with Wright-Giemsa stain (center, WG) and toluidine blue (right, TB). (C) The ratio of 4-OHT (+) relative to 4-OHT (−) BMMCs prepared from CreERT2, ΔCF/+, and ΔCF/ΔCF mice. The cells were counted on days 3 and 6 of the 4-OHT treatment (D3 and D6, respectively). (D) The percentages of dead cells in the CreERT2 and ΔCF/ΔCF BMMCs cultured in the absence (−) or presence (+) of 4-OHT for 3 and 6 days (D3 and D6, respectively). In C and D, no cells survived in the absence of IL-3 and SCF [cytokine(−)]. (E) (Left) Representative FACS data for the 5-bromo-2'-deoxyuridine and 7-amino-actinomycin D expression of ΔCF/ΔCF BMMCs on days 0, 4, and 8 of 4-OHT treatment (D0, D4, and D8, respectively). The lower right panel shows the gates displaying the cell cycle populations in the subG0/G1 (Sub), G0/G1, S, and G2/M phases. (Right) Average percentages and SD of the gated cells. *P < .05, **P < 0.01. N = 4. (F) The results of a FACS analysis of the presorted c-Kit/FcεRIα DP BMMCs from Gata2flox/flox::Rosa26CreERT2 mice after 0, 3, 6, and 9 days of 4-OHT treatment (D0, D3, D6, and D9, respectively). The average percentages and SD of the gated cells are indicated. N = 4.
The deletion of GATA2 CF results in a loss of mast cell identity in mouse bone marrow-derived mast cells. (A) (Left) Representative FACS data for BMMCs prepared from Gata2flox/flox::Rosa26CreERT2 mice after 0, 3, 6, and 9 days of 4-OHT treatment (D0, D3, D6, and D9, respectively). (Right) Average percentages and SD of the DP cells within the gates examined on days 0, 3, 5, 7, and 9 of the 4-OHT treatment (D0, D3, D5, D7, and D9, respectively). N = 9. (B) Representative bright field (BF) phase contrast photomicrographs (left; original magnification, ×200) and cytospin preparations (center and right; original magnification, ×400) of BMMCs prepared from Gata2+/+::Rosa26CreERT2 (CreERT2) and Gata2flox/flox::Rosa26CreERT2(ΔCF/ΔCF) mice treated with 0.5 μM of 4-OHT for 10 days. For cytospin preparations, the cells were stained with Wright-Giemsa stain (center, WG) and toluidine blue (right, TB). (C) The ratio of 4-OHT (+) relative to 4-OHT (−) BMMCs prepared from CreERT2, ΔCF/+, and ΔCF/ΔCF mice. The cells were counted on days 3 and 6 of the 4-OHT treatment (D3 and D6, respectively). (D) The percentages of dead cells in the CreERT2 and ΔCF/ΔCF BMMCs cultured in the absence (−) or presence (+) of 4-OHT for 3 and 6 days (D3 and D6, respectively). In C and D, no cells survived in the absence of IL-3 and SCF [cytokine(−)]. (E) (Left) Representative FACS data for the 5-bromo-2'-deoxyuridine and 7-amino-actinomycin D expression of ΔCF/ΔCF BMMCs on days 0, 4, and 8 of 4-OHT treatment (D0, D4, and D8, respectively). The lower right panel shows the gates displaying the cell cycle populations in the subG0/G1 (Sub), G0/G1, S, and G2/M phases. (Right) Average percentages and SD of the gated cells. *P < .05, **P < 0.01. N = 4. (F) The results of a FACS analysis of the presorted c-Kit/FcεRIα DP BMMCs from Gata2flox/flox::Rosa26CreERT2 mice after 0, 3, 6, and 9 days of 4-OHT treatment (D0, D3, D6, and D9, respectively). The average percentages and SD of the gated cells are indicated. N = 4.
GATA2-deficient BMMCs show characteristics of immature myeloid-like cells
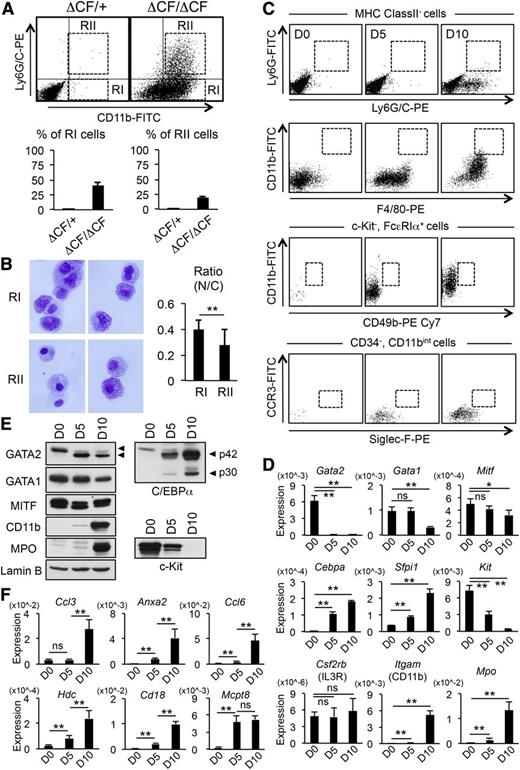
We next characterized the ΔCF/ΔCF BMMCs by examining the expression of myeloid cell markers by FACS (Figure 2). Notably, the expression of CD11b, a marker for monocyte and granulocyte lineages, was significantly increased in ΔCF/ΔCF BMMCs (Figure 2A). Furthermore, some of the CD11b-positive cells were also positive for Ly6G/C. Both the Ly6G/C−CD11b+ (RI) and Ly6G/C+CD11b+ (RII) fractions were composed of cells showing heterogeneous morphological features on the cytospin preparations stained with Wright-Giemsa (Figure 2B). The average N/C ratio was lower in RII than in RI, suggesting that there was a greater number of mature cells in the RII fraction (Figure 2B). Next, we examined whether a subset of ΔCF/ΔCF cells expresses the markers of specific cell lineages (Figure 2C). In addition to Ly6G/C and CD11b, moderate increases in Siglec-F and F4/80 expression were observed in a subset of ΔCF/ΔCF cells. However, on the basis of the combination of 2 or more cell lineage markers, neutrophils18 (MHCclassII−Ly6G+Ly6G/C+), monocytes/macrophages,19-21 (CD11bhighF4/80+), basophils (c-Kit−FcεRIα+CD11bintCD49b+), and eosinophils (CD34−CD11bintCCR3+Siglec-F+) were not detected on days 5 and 10 following 4-OHT treatment. In accordance with these results, the Q-PCR analyses revealed a significant reduction of the mRNA level of c-Kit and an increased level of CD11b (Itgam) mRNA (Figure 2D). In contrast, the IL-3RB (Csf2rb) mRNA level was not affected by the GATA2 CF deletion. We also observed an increased mRNA level of the 2 myeloid cell transcription factors, C/EBPα (Cebpa) and PU.1 (Sfpi1),12-14 and myeloperoxidase (Mpo), which is known to be regulated by C/EBPα.22 On the other hand, the levels of 2 other transcription factors expressed in mast cells, GATA1 and MITF, were gradually decreased by day 10. The abundance of the corresponding proteins was changed similarly to the changes in the mRNA as determined by a western blot analysis (Figure 2E). The upregulation of C/EBPα mRNA was also observed in wild-type BMMCs treated with siRNA against GATA2, but not GATA1, excluding a dominant negative effect by the GATA2ΔCF protein. Furthermore, this upregulation was partly rescued by transducing the cells with GATA2 cDNA, suggesting that the upregulation of Cebpa was indeed a consequence of disrupting GATA2 function (supplemental Figure 2).
The GATA2-deficient BMMCs show characteristics of immature myeloid-like cells. (A) (Upper) BMMCs prepared from ΔCF/+ and ΔCF/ΔCF mice were stained with CD11b and Ly6G/C antibodies and analyzed by FACS after 10 days of 4-OHT treatment. The CD11b+/Ly6G/C− and CD11b+/Ly6G/C+ cells were gated as RI and RII fractions, respectively. (Lower) Average percentages and SD of the RI (left) fractions in the ΔCF/+ (0.2 ± 0.2%) and ΔCF/ΔCF BMMCs (40.7 ± 5.8%) and RII (right) fractions in the ΔCF/+ (0.3 ± 0.1%) and ΔCF/ΔCF(19.7 % ± 1.5%) BMMCs are shown. N = 4. (B) (Left) Representative cytospin preparations of the RI (upper) and RII (lower) cells stained with Wright-Giemsa. Original magnification, ×400. (Right) Mean values of N/C ratio of the RI and RII cells. N = 30. (C) Representative flow cytometric plots of neutrophils (MHCclassII−Ly6G+Ly6G/C+), monocytes (CD11bhighF4/80+), basophils (c-Kit−FcεRIα+CD11bintCD49b+), and eosinophils (CD34−CD11bintCCR3+Siglec-F+) from ΔCF/ΔCF BMMCs after 0, 5, and 10 days of 4-OHT treatment. (D) The results of the Q-PCR analyses of the indicated genes in ΔCF/ΔCF BMMCs after 0, 5, and 10 days of 4-OHT treatment. N = 4. (E) The expression of the GATA2, GATA1, MITF, CD11b, MPO, C/EBPα, c-Kit, and Lamin-B (control) proteins was analyzed by western blot analysis. Whole cell lysates were isolated from ΔCF/ΔCF BMMCs treated with 4-OHT for 0, 5, and 10 days. The arrowheads indicate the positions of the wild-type and ΔCF GATA2 protein and the p42 and p30 isoforms of C/EBPα. (F) The results of the Q-PCR analyses of basophil-enriched genes in the ΔCF/ΔCF BMMCs after 0, 5, and 10 days of 4-OHT treatment. N = 4. For B, D, and F. *P < .05, **P < .01, ns, not significant. For C-F, samples prepared after 0, 5, and 10 days of 4-OHT treatment are indicated by D0, D5, and D10, respectively.
The GATA2-deficient BMMCs show characteristics of immature myeloid-like cells. (A) (Upper) BMMCs prepared from ΔCF/+ and ΔCF/ΔCF mice were stained with CD11b and Ly6G/C antibodies and analyzed by FACS after 10 days of 4-OHT treatment. The CD11b+/Ly6G/C− and CD11b+/Ly6G/C+ cells were gated as RI and RII fractions, respectively. (Lower) Average percentages and SD of the RI (left) fractions in the ΔCF/+ (0.2 ± 0.2%) and ΔCF/ΔCF BMMCs (40.7 ± 5.8%) and RII (right) fractions in the ΔCF/+ (0.3 ± 0.1%) and ΔCF/ΔCF(19.7 % ± 1.5%) BMMCs are shown. N = 4. (B) (Left) Representative cytospin preparations of the RI (upper) and RII (lower) cells stained with Wright-Giemsa. Original magnification, ×400. (Right) Mean values of N/C ratio of the RI and RII cells. N = 30. (C) Representative flow cytometric plots of neutrophils (MHCclassII−Ly6G+Ly6G/C+), monocytes (CD11bhighF4/80+), basophils (c-Kit−FcεRIα+CD11bintCD49b+), and eosinophils (CD34−CD11bintCCR3+Siglec-F+) from ΔCF/ΔCF BMMCs after 0, 5, and 10 days of 4-OHT treatment. (D) The results of the Q-PCR analyses of the indicated genes in ΔCF/ΔCF BMMCs after 0, 5, and 10 days of 4-OHT treatment. N = 4. (E) The expression of the GATA2, GATA1, MITF, CD11b, MPO, C/EBPα, c-Kit, and Lamin-B (control) proteins was analyzed by western blot analysis. Whole cell lysates were isolated from ΔCF/ΔCF BMMCs treated with 4-OHT for 0, 5, and 10 days. The arrowheads indicate the positions of the wild-type and ΔCF GATA2 protein and the p42 and p30 isoforms of C/EBPα. (F) The results of the Q-PCR analyses of basophil-enriched genes in the ΔCF/ΔCF BMMCs after 0, 5, and 10 days of 4-OHT treatment. N = 4. For B, D, and F. *P < .05, **P < .01, ns, not significant. For C-F, samples prepared after 0, 5, and 10 days of 4-OHT treatment are indicated by D0, D5, and D10, respectively.
Previous studies showed that C/EBPα plays a critical role in specifying the basophil cell fate.23-25 Although the ΔCF/ΔCF cells were negative for CD49b, a basophil marker, Q-PCR showed that there was a significant increase in the expression of basophil-enriched genes regulated by C/EBPα (Figure 2F).24 Furthermore, the mRNA level of Mcpt8, another lineage-specific marker for mouse basophils,26 was significantly increased by day 5 (Figure 2F). These results indicate that the ΔCF/ΔCF cells could not be classified into any of the specific cell lineages on the basis of cell surface markers. Importantly, however, these cells showed some features of basophils on the basis of their gene expression profiles.
GATA2-deficient BMMCs give rise to neutrophil- and macrophage-like cells, but not to eosinophils, in the presence of myeloid cytokines
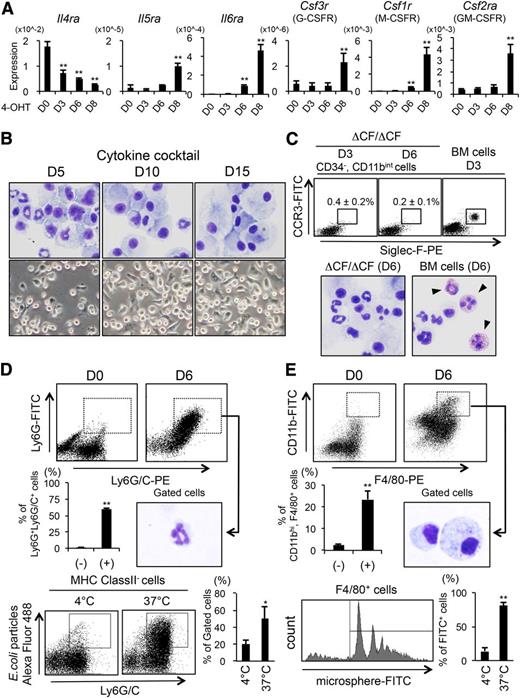
We next examined the mRNA expression of receptors for myeloid cytokines in the ΔCF/ΔCF BMMCs on days 0, 3, 6, and 8 of 4-OHT treatment by Q-PCR (Figure 3A). The receptor for IL-4, which is known to be expressed in mast cells,27 was significantly downregulated in the ΔCF/ΔCF cells. In contrast, the expression of a variety of genes encoding other cytokine receptors, including Il5ra, Il6ra, Csf3r, Csf1r, and Csf2ra, was upregulated. These findings raise the possibility that the ΔCF/ΔCF cells possess the potential to differentiate into other myeloid lineage cells in the presence of these cytokines. To test this possibility, the ΔCF/ΔCF cells were treated with 4-OHT for 10 days and then were cultured for an additional 15 days with a cytokine cocktail consisting of IL-3, granulocyte colony-stimulating factor (CSF), macrophage-CSF, granulocyte macrophage–CSF, IL-5, and IL-6 (Figure 3B; supplemental Figure 3A). We observed that, although most of the cells were nonadherent on day 5 of culture, the number of adherent cells was increased over time, and these cells became dominant by day 15. Cytospin preparations of nonadherent cells stained with Diff-Quik showed that the ΔCF/ΔCF cells were composed of a heterogeneous cell population including macrophage-like mononuclear cells and neutrophil-like cells featured with multilobed nuclei. The macrophage-like cells became predominant over time, and the cytoplasm of these cells was enlarged by day 15. The FACS analyses showed that neutrophils (MHC classII−Ly6G+Ly6G/C+) appeared in response to the cytokine cocktail (supplemental Figure 3A). The monocyte/macrophage fraction (CD11bhighMHCClassII+F4/80+CD11c+) was markedly expanded from days 10 to 15 of culture, and these cells made up almost all of the adherent cell fraction. On the other hand, basophils (c-Kit−FcεRIα+CD11bintCD49b+) and eosinophils (CD34−CD11bintCCR3+Siglec-F+) were rarely observed at any time points under this culture condition (supplemental Figure 3A).
The GATA2-deficient BMMCs are able to differentiate into functional neutrophil- and macrophage-like cells, but not into eosinophils, in the presence of myeloid cytokines. (A) The results of the Q-PCR analyses of receptors of the myeloid linage in ΔCF/ΔCF BMMCs. The PCR products amplified from ΔCF/ΔCF BMMCs on days 0, 3, 6, and 8 of the 4-OHT treatment (D0, D3, D6, and D8, respectively) are shown. N = 4. (B) Representative cytospin preparations stained with Wright-Giemsa (upper; original magnification, ×400) and phase-contrast photomicrographs (lower; original magnification, ×200) of ΔCF/ΔCF BMMCs cultured with the cytokine cocktail for 5, 10, and 15 days following the 10 days of 4-OHT treatment (D0, D5, and D10, respectively). (C) (Upper) Results of a FACS analysis for eosinophils (CD34−CD11bintCCR3+Siglec-F+) from ΔCF/ΔCF BMMCs cultured with the culture media appropriate for eosinophils containing 20 ng/mL of IL-5 (Supplemental methods) for 3 and 6 days (D3 and D6, respectively) following 10 days of 4-OHT treatment. The average percentages and SD of the gated cells are indicated. N = 3. The results of the FACS analysis of bone marrow cells from wild-type mice (BM cells) cultured for 3 days under the same culture conditions are shown in the right panel. (Lower) Cytospin preparations stained with Diff-quick from the ΔCF/ΔCF BMMCs and BM cells cultured for 6 days under the conditions described above. (D) (Top) Results of a FACS analyses of neutrophil markers on ΔCF/ΔCF BMMCs cultured under the conditions for neutrophil amplification for 6 days. (Middle) Average percentages and SD of the gated fractions (left); cytospin preparations of the isolated Ly6G+Ly6G/C+ neutrophil-like cells stained with Wright-Giemsa (right). (Bottom, left) Results of the FACS analyses of the Alexa Fluor 488-conjugated E coli bioparticles phagocytosed by the MHC class II− and Ly6G/C+ cells cultured at 4°C and 37°C. (Right) Average percentages and SD of the gated fractions. The representative data from 3 independent analyses are shown. (E) (Top) Results of the FACS analyses of macrophage markers (CD11b and F4/80) on ΔCF/ΔCF BMMCs cultured under the conditions for macrophage amplification for 6 days. (Middle) Average percentages and SD of the gated fractions (left); cytospin preparations of the isolated CD11b+F4/80+ macrophage-like cells stained with Wright-Giemsa (right). (Bottom, left) FACS histogram of microspheres phagocytosed by the F4/80+ cells cultured at 37°C. (Right) Average percentages and SD of the phagocytosed cells cultured at 4°C and 37°C. The representative data from 4 independent analyses are shown. Original magnification, ×400 for C-E.
The GATA2-deficient BMMCs are able to differentiate into functional neutrophil- and macrophage-like cells, but not into eosinophils, in the presence of myeloid cytokines. (A) The results of the Q-PCR analyses of receptors of the myeloid linage in ΔCF/ΔCF BMMCs. The PCR products amplified from ΔCF/ΔCF BMMCs on days 0, 3, 6, and 8 of the 4-OHT treatment (D0, D3, D6, and D8, respectively) are shown. N = 4. (B) Representative cytospin preparations stained with Wright-Giemsa (upper; original magnification, ×400) and phase-contrast photomicrographs (lower; original magnification, ×200) of ΔCF/ΔCF BMMCs cultured with the cytokine cocktail for 5, 10, and 15 days following the 10 days of 4-OHT treatment (D0, D5, and D10, respectively). (C) (Upper) Results of a FACS analysis for eosinophils (CD34−CD11bintCCR3+Siglec-F+) from ΔCF/ΔCF BMMCs cultured with the culture media appropriate for eosinophils containing 20 ng/mL of IL-5 (Supplemental methods) for 3 and 6 days (D3 and D6, respectively) following 10 days of 4-OHT treatment. The average percentages and SD of the gated cells are indicated. N = 3. The results of the FACS analysis of bone marrow cells from wild-type mice (BM cells) cultured for 3 days under the same culture conditions are shown in the right panel. (Lower) Cytospin preparations stained with Diff-quick from the ΔCF/ΔCF BMMCs and BM cells cultured for 6 days under the conditions described above. (D) (Top) Results of a FACS analyses of neutrophil markers on ΔCF/ΔCF BMMCs cultured under the conditions for neutrophil amplification for 6 days. (Middle) Average percentages and SD of the gated fractions (left); cytospin preparations of the isolated Ly6G+Ly6G/C+ neutrophil-like cells stained with Wright-Giemsa (right). (Bottom, left) Results of the FACS analyses of the Alexa Fluor 488-conjugated E coli bioparticles phagocytosed by the MHC class II− and Ly6G/C+ cells cultured at 4°C and 37°C. (Right) Average percentages and SD of the gated fractions. The representative data from 3 independent analyses are shown. (E) (Top) Results of the FACS analyses of macrophage markers (CD11b and F4/80) on ΔCF/ΔCF BMMCs cultured under the conditions for macrophage amplification for 6 days. (Middle) Average percentages and SD of the gated fractions (left); cytospin preparations of the isolated CD11b+F4/80+ macrophage-like cells stained with Wright-Giemsa (right). (Bottom, left) FACS histogram of microspheres phagocytosed by the F4/80+ cells cultured at 37°C. (Right) Average percentages and SD of the phagocytosed cells cultured at 4°C and 37°C. The representative data from 4 independent analyses are shown. Original magnification, ×400 for C-E.
Because the ΔCF/ΔCF cells were weakly positive for an eosinophil marker, Siglec-F+, the capacity to develop into this lineage was further assessed under the culture conditions appropriate for the bone marrow-derived eosinophils (Figure 3C). Unexpectedly, the CD34−CD11bintCCR3+Siglec-F+ eosinophil fraction never differentiated from the ΔCF/ΔCF cells, ruling out the possibility that the cells have the potential to develop into this cell lineage. Consistent with these results, the expression of eosinophil-specific genes (Il5ra, Cebpb, and Epx) was unchanged under this culture condition (supplemental Figure 3B). We then cultured the ΔCF/ΔCF cells under culture conditions for the specific amplification of neutrophils or macrophages (Figure 3D-E).28-31 Following a 6-day culture under these conditions, these cells were matured into Ly6G+Ly6G/C+ neutrophils (59 ± 3.7%) or CD11bhiF4/80+ macrophages (23 ± 3.8%) (Figure 3D-E, top). The Ly6G+Ly6G/C+ and CD11bhiF4/80+ gated cells showed morphological features of neutrophils and macrophages, respectively (Figure 3D-E, middle). Furthermore, both neutrophil- and macrophage-like cells showed phagocytic activity against fluorescently labeled bioparticles (Figure 3D-E, bottom). Collectively, these results indicate that the ΔCF/ΔCF cells are able to differentiate into functional macrophage- and neutrophil-like cells in the presence of myeloid cytokines.
Genome-wide gene expression analysis revealed downregulation of mast cell-specific genes and upregulation of myeloid genes in GATA2-deficient BMMCs
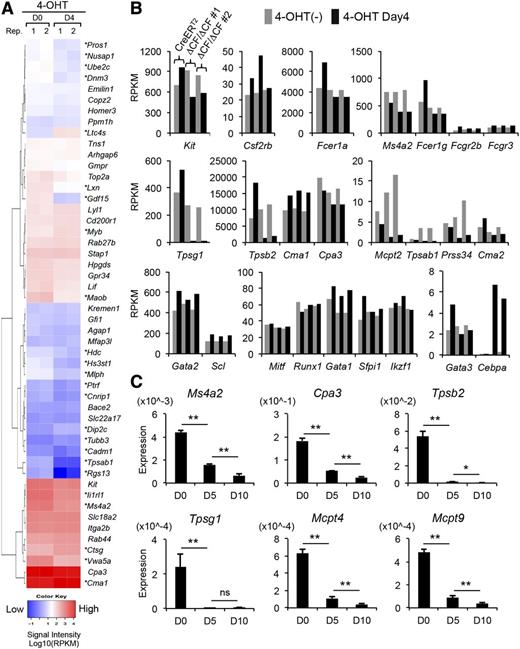
To gain insight into the roles of GATA2 in mast cells, we comprehensively examined the gene expression profiles in the ΔCF/ΔCF cells by performing RNA-Seq and subsequent gene ontology (GO) analyses. No significant enrichment of any GO terms (false discovery rate < 0.01) was found among the 169 downregulated genes. On the other hand, among the 217 upregulated genes, the GO biological process categories related to metabolic processes and processing of RNA and the corresponding cellular component categories were significantly overrepresented (supplemental Table 1). However, no significant enrichment of molecular function categories was observed in these genes. Thus, further analyses are needed to define the biological significance of the upregulated genes. In a recent comprehensive study of the human mast cell transcriptome using cap analysis of gene expression technology, 50 mast cell-specific genes were demonstrated to be coexpressed with GATA1 and GATA2 to form a gene cluster.32 We found that the expression of 24 mouse homologs of these 50 cluster genes was significantly affected by the GATA2 deletion (18 downregulated and 6 upregulated genes; asterisks in Figure 4A). We also found that the expression of several well-defined mast cell marker genes was decreased by the GATA2 CF deletion (Figure 4B, top and middle). Notably, the expression levels of mast cell transcription factors, such as Scl, Mitf, and Sfpi1, were rarely affected, whereas the expression of Cebpa was most significantly increased by the GATA2 ablation (Figure 4B, bottom). The downregulation of several mast cell-specific genes was also observed on days 5 and 10 of 4-OHT treatment, as detected by Q-PCR (Figure 4C). Collectively, these data suggest that GATA2 plays an essential role in the regulation of mast cell-specific genes in BMMCs.
Genome-wide transcriptome profiling revealed a wide influence of mast cell-specific gene expression in the GATA2-deficient BMMCs. (A) Heat map showing color-coded expression levels of GATA factor-related genes in the GATA2-deficeint BMMCs. The RPKM values were obtained from 2 ΔCF/ΔCF BMMCs samples. Red indicates relatively high expression; blue indicates low expression; and white indicates intermediate expression in 4-OHT-treated (D4) vs untreated ΔCF/ΔCF BMMCs (D0), as indicated by the color bar. *Differentially expressed genes with a fold-change (D4/D0) <0.5 or >1.5. (B) The RPKM values of the RNA-seq samples prepared from 4-OHT-treated (black bars) and untreated (gray bars) BMMCs. (C) The results of the Q-PCR analyses of mast cell-specific genes in the ΔCF/ΔCF BMMCs. The PCR products amplified from ΔCF/ΔCF BMMCs on days 0, 5, and 10 of the 4-OHT treatment (D0, D5, and D10, respectively) are shown. N = 4.
Genome-wide transcriptome profiling revealed a wide influence of mast cell-specific gene expression in the GATA2-deficient BMMCs. (A) Heat map showing color-coded expression levels of GATA factor-related genes in the GATA2-deficeint BMMCs. The RPKM values were obtained from 2 ΔCF/ΔCF BMMCs samples. Red indicates relatively high expression; blue indicates low expression; and white indicates intermediate expression in 4-OHT-treated (D4) vs untreated ΔCF/ΔCF BMMCs (D0), as indicated by the color bar. *Differentially expressed genes with a fold-change (D4/D0) <0.5 or >1.5. (B) The RPKM values of the RNA-seq samples prepared from 4-OHT-treated (black bars) and untreated (gray bars) BMMCs. (C) The results of the Q-PCR analyses of mast cell-specific genes in the ΔCF/ΔCF BMMCs. The PCR products amplified from ΔCF/ΔCF BMMCs on days 0, 5, and 10 of the 4-OHT treatment (D0, D5, and D10, respectively) are shown. N = 4.
Increased expression of C/EBPα is essential for the dedifferentiation of GATA2-deficient BMMCs
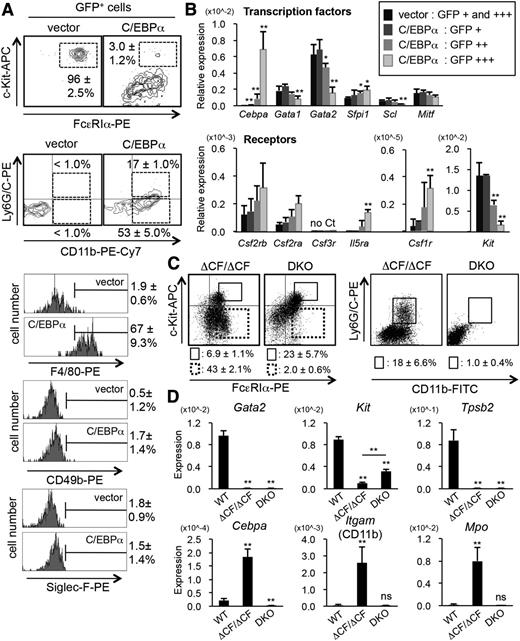
Next, we examined whether the phenotype of the ΔCF/ΔCF BMMCs was recapitulated by retroviral expression of C/EBPα in wild-type BMMCs (Figure 5). We found that the number of DP cells was reduced, whereas the expression of CD11b, Ly6G/C, and F4/80 was increased in the C/EBPα-expressing cells (Figure 5A). The expression of CD49b and Siglec-F was not increased in these cells. The Q-PCR analyses showed that the mRNA levels of c-Kit were reduced, whereas those of the cytokine receptors were dependent on the expression level of C/EBPα, except for Csf3r (Figure 5B). Interestingly, the endogenous GATA2 mRNA levels were inversely correlated with those of C/EBPα, suggesting the existence of a crossregulation between C/EBPα and GATA2 in BMMCs.
Forced expression of C/EBPα in BMMCs results in the dedifferentiation of BMMCs, recapitulating the characteristics of the ΔCF/ΔCF cells. (A) The results of the FACS analyses of mast cell (c-Kit and FcεRIα), myeloid cell (Ly6G/C and CD11b), macrophage (F4/80), basophil (CD49b), and eosinophil (Siglec-F) markers on BMMCs retrovirally transduced with either a control vector pMXs IG40,41 or vector expressing C/EBPα cDNA by using a retroviral packaging cell line, PlatE.42 Representative data from ≥5 independent analyses are shown. (B) The results of the Q-PCR analyses of transcription factors (upper) and cytokine receptors (lower) in the C/EBPα-expressing and control BMMCs. BMMCs were transfected with the pBAT12 IRES eGFP vector or pBAT12 Cebpa IRES eGFP by electroporation. The cell populations were sorted based on the intensity of GFP expression, as indicated. *P < .05; **P < .01 (compared with data from the vector-transfected cells). N = 4. (C) The results of the FACS analyses of mast cells (c-Kit and FcεRIα) and myeloid cells (Ly6G/C and CD11b) from ΔCF/ΔCF and ΔCF/ΔCF::Cebpa−/− DKO BMMCs. The BMMCs were treated with 4-OHT for 9 days. The average percentages and SD of the DP and SP cells within the gates are shown. N = 4. (D) The results of the Q-PCR analyses of the indicated genes in wild-type, ΔCF/ΔCF, and DKO BMMCs after 9 days of 4-OHT treatment. *P < .05; **P < .01; ns, not significant (compared with data from wild-type cells). N = 4.
Forced expression of C/EBPα in BMMCs results in the dedifferentiation of BMMCs, recapitulating the characteristics of the ΔCF/ΔCF cells. (A) The results of the FACS analyses of mast cell (c-Kit and FcεRIα), myeloid cell (Ly6G/C and CD11b), macrophage (F4/80), basophil (CD49b), and eosinophil (Siglec-F) markers on BMMCs retrovirally transduced with either a control vector pMXs IG40,41 or vector expressing C/EBPα cDNA by using a retroviral packaging cell line, PlatE.42 Representative data from ≥5 independent analyses are shown. (B) The results of the Q-PCR analyses of transcription factors (upper) and cytokine receptors (lower) in the C/EBPα-expressing and control BMMCs. BMMCs were transfected with the pBAT12 IRES eGFP vector or pBAT12 Cebpa IRES eGFP by electroporation. The cell populations were sorted based on the intensity of GFP expression, as indicated. *P < .05; **P < .01 (compared with data from the vector-transfected cells). N = 4. (C) The results of the FACS analyses of mast cells (c-Kit and FcεRIα) and myeloid cells (Ly6G/C and CD11b) from ΔCF/ΔCF and ΔCF/ΔCF::Cebpa−/− DKO BMMCs. The BMMCs were treated with 4-OHT for 9 days. The average percentages and SD of the DP and SP cells within the gates are shown. N = 4. (D) The results of the Q-PCR analyses of the indicated genes in wild-type, ΔCF/ΔCF, and DKO BMMCs after 9 days of 4-OHT treatment. *P < .05; **P < .01; ns, not significant (compared with data from wild-type cells). N = 4.
We then examined whether the phenotype of the ΔCF/ΔCF BMMCs was rescued by simultaneous disruption of the Cebpa and Gata2 genes using conditional double-knockout (DKO) mice (Figure 5C). Surprisingly, the reduction of DP cells was still observed, whereas the emergence of the Ly6G/C+CD11b+ cells vanished after the simultaneous deletion of the Cebpa and Gata2 genes in the ΔCF/ΔCF BMMCs. Intriguingly, within the c-Kit− cell fraction, the number of FcεRIα+ single-positive (SP) cells was significantly reduced in the DKO cells compared with the ΔCF/ΔCF cells (2.0 ± 0.6% and 43.0 ± 2.1%, respectively). These data suggest that the expression of FcεRIα+ is dependent on C/EBPα in the ΔCF/ΔCF cells. The Q-PCR analyses revealed that the mRNA levels of CD11b and Mpo were dependent on C/EBPα, whereas those of Kit and Mcpt6 were reduced by the loss of GATA2, irrespective of the C/EBPα level (Figure 5D).
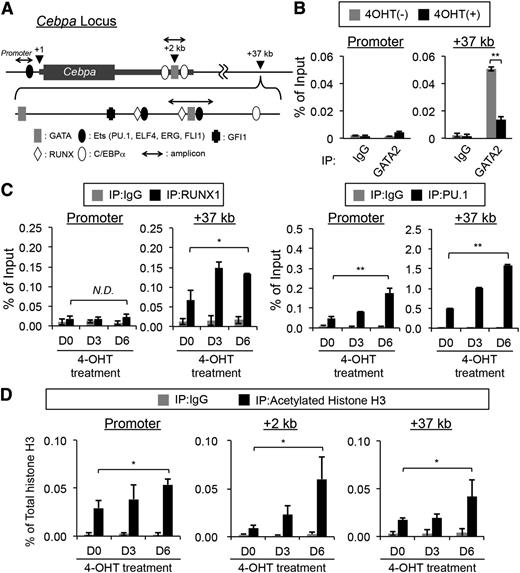
GATA2 binds to the +37-kb enhancer chromatin region of the murine Cebpa locus
To examine whether GATA2 directly regulates the expression of the Cebpa gene, putative GATA2 binding sequences were searched on the Cebpa locus (Figure 6A). The conserved GATA binding sites were found in the 2 cis-acting regions, referred to as +2 and +37 kb, and the latter has been reported as an enhancer activated by RUNX1 in an immature myeloid cell line, 32Dcl3.33-35 Interestingly, there is a 34-bp region containing the GATA, RUNX1, and PU.1 binding sites in close proximity in this region (Figure 6A; supplemental Figure 4). GATA2 binding to this region, but not to the promoter, was detected in BMMCs, and this binding activity was significantly reduced after 4-OHT treatment (Figure 6B). Furthermore, the binding of RUNX1 and PU.1 to this region was significantly increased by GATA2 CF deletion (Figure 6C). These data suggest that the binding of GATA2 attenuates the binding of RUNX1 and PU.1 to the +37-kb chromatin region. Interestingly, the GATA2 CF deletion increased the acetylation of histone H3 not only at the +37-kb region, but also at the promoter and +2-kb regions (Figure 6D). Overall, the increased Cebpa expression in the ΔCF/ΔCF BMMCs could be explained, at least in part, by the recruitment of RUNX1 and PU.1 to the +37-kb region in the absence of GATA2.
GATA2 binding is enriched in the +37-kb chromatin region of the Cebpa gene. (A) The configuration of the murine Cebpa gene. The consensus DNA-binding sequences for transcription factors are shown as indicated. The positions of the PCR amplicons used in the qChIP assays are indicated by 3-headed arrows. (B) The DNA-binding activity of GATA2 to the Cebpa promoter and +37-kb region in ΔCF/ΔCF BMMCs was examined using qChIP assays. Black and gray bars indicate 4-OHT-treated (for 3 days) and untreated ΔCF/ΔCF BMMCs, respectively. N = 4. The control experiments were performed using rabbit IgG in place of anti-GATA2 antibodies. (C) qChIP assays of the Cebpa promoter and +37-kb regions were performed with anti RUNX1 (left 2 panels) and PU.1 (right 2 panels) antibodies. (D) qChIP assays of the Cebpa promoter and +2- and +37-kb regions were performed with antiacetylated histone H3 antibodies. In C and D, the samples were prepared from ΔCF/ΔCF BMMCs treated with 4-OHT for 0, 3, and 6 days (D0, D3, and D6). Gray bars indicate the control experiments using rabbit IgG in place of the indicated antibodies. *P < .05; **P < .01 (compared with the data for D0). N = 4.
GATA2 binding is enriched in the +37-kb chromatin region of the Cebpa gene. (A) The configuration of the murine Cebpa gene. The consensus DNA-binding sequences for transcription factors are shown as indicated. The positions of the PCR amplicons used in the qChIP assays are indicated by 3-headed arrows. (B) The DNA-binding activity of GATA2 to the Cebpa promoter and +37-kb region in ΔCF/ΔCF BMMCs was examined using qChIP assays. Black and gray bars indicate 4-OHT-treated (for 3 days) and untreated ΔCF/ΔCF BMMCs, respectively. N = 4. The control experiments were performed using rabbit IgG in place of anti-GATA2 antibodies. (C) qChIP assays of the Cebpa promoter and +37-kb regions were performed with anti RUNX1 (left 2 panels) and PU.1 (right 2 panels) antibodies. (D) qChIP assays of the Cebpa promoter and +2- and +37-kb regions were performed with antiacetylated histone H3 antibodies. In C and D, the samples were prepared from ΔCF/ΔCF BMMCs treated with 4-OHT for 0, 3, and 6 days (D0, D3, and D6). Gray bars indicate the control experiments using rabbit IgG in place of the indicated antibodies. *P < .05; **P < .01 (compared with the data for D0). N = 4.
Cebpa expression was not increased in the ΔCF/ΔCF peritoneal mast cells
Finally, we investigated whether our findings in BMMCs were also observed in cultured mast cells derived from the peritoneal cavity (PMCs). As observed in BMMCs, the DP cell fraction was clearly reduced on day 9 following 4-OHT treatment, although the impact was smaller than that in BMMCs (Figure 7A). Interestingly, FcεRIα SP cells were rarely observed, similar to our finding in the DKO BMMCs (Figure 5C). Moreover, unlike in the ΔCF/ΔCF BMMCs, the CD11bint cells did not appear in the ΔCF/ΔCF PMCs. These findings led us to predict that C/EBPα mRNA may not be upregulated in the ΔCF/ΔCF PMCs. Indeed, the mRNA levels of Cebpa, Mpo, and CD11b were not increased in the ΔCF/ΔCF PMCs. In contrast, reduced c-Kit expression on the cell surface and reduced Mcpt6 and Tpsg1 mRNA levels were consistently observed in both BMMCs and PMCs, irrespective of the C/EBPα level (Figure 7B).
No upregulation of Cebpa is observed in ΔCF/ΔCF peritoneal mast cells. (A) Representative FACS data for markers of mast cells (c-Kit and FcεRIα) and basophils (CD11b and CD49b) on ΔCF/ΔCF PMCs. PMCs were cultured ex vivo for 14 days and then were cultured in the absence (−) or presence (+) of 4-OHT for an additional 9 days. The average percentages and SD of the DP cells within the gates are shown. N = 4. (B) The results of the Q-PCR analyses of the indicated genes in PMCs and BMMCs prepared from wild-type mice (WT) and ΔCF/ΔCF PMCs cultured in the absence (−) or presence (+) of 4-OHT. *P < .05; **P < .01 [compared with data from 4-OHT(−) ΔCF/ΔCF PMCs]. N = 4. (C-D) Schematic summaries of the present study. (C) Summary of the reprogramming activity of ΔCF/ΔCF BMMCs. (D) Summary of the phenotypes of the genetically manipulated BMMCs and PMCs used in this study.
No upregulation of Cebpa is observed in ΔCF/ΔCF peritoneal mast cells. (A) Representative FACS data for markers of mast cells (c-Kit and FcεRIα) and basophils (CD11b and CD49b) on ΔCF/ΔCF PMCs. PMCs were cultured ex vivo for 14 days and then were cultured in the absence (−) or presence (+) of 4-OHT for an additional 9 days. The average percentages and SD of the DP cells within the gates are shown. N = 4. (B) The results of the Q-PCR analyses of the indicated genes in PMCs and BMMCs prepared from wild-type mice (WT) and ΔCF/ΔCF PMCs cultured in the absence (−) or presence (+) of 4-OHT. *P < .05; **P < .01 [compared with data from 4-OHT(−) ΔCF/ΔCF PMCs]. N = 4. (C-D) Schematic summaries of the present study. (C) Summary of the reprogramming activity of ΔCF/ΔCF BMMCs. (D) Summary of the phenotypes of the genetically manipulated BMMCs and PMCs used in this study.
Discussion
In this study, we examined the roles of GATA2 in mast cell development by using BMMCs and PMCs. There were 2 major findings obtained from these studies. The first is about the reprogramming of the ΔCF/ΔCF BMMCs into other cell lineages (Figure 7C). These cells showed the potential to differentiate into macrophage- and neutrophil-like cells, but not into eosinophils, under the appropriate culture conditions. Interestingly, the Q-PCR analyses showed that there was increased expression of several basophil-enriched genes. Moreover, a subpopulation of the ΔCF/ΔCF cells was c-Kit−FcεRIα+CD11bint, suggesting that they had characteristics of basophils. However, CD49b, a representative marker for basophils, never appeared on these cells under any culture conditions. In addition, the expression of MITF, which is reported to be absent in basophils,24 was not silenced in the ΔCF/ΔCF cells. Thus, we concluded that the differentiation of these cells into basophils was aberrant or incomplete in the absence of GATA2. These findings are related to an earlier study reported by Iwasaki et al, which showed the importance of the order of GATA2 and C/EBPα activation for the lineage specification of eosinophils, basophils, and mast cells.25 The current study extends these findings by demonstrating the requirement of GATA2 for the differentiation of these cells into these various cell lineages. Our results showed that GATA2 is absolutely and partially required for eosinophil and basophil development, respectively, whereas the contribution of this factor is much lower in neutrophil and macrophage development (Figure 7C).
The second major finding is related to the cross-antagonism between GATA2 and C/EBPα (Figure 7D). Recently, Qi et al reported that C/EBPα and MITF repress each other’s transcription in “pre-BMPs,” a subset of granulocyte-monocyte progenitors. These cells mostly give rise to basophils and mast cells while retaining a limited capacity to differentiate into other myeloid cells.24 In a separate study, another myeloid and lymphoid transcription factor, Ikaros (encoded by Ikzf1), and its downstream factor, Hes1, were found to epigenetically downregulate Cebpa gene expression and to thereby limit the differentiation of basophils.34 These studies, together with our findings, indicate that the Cebpa gene can be repressed by several molecular mechanisms. Although the origin and developmental pathways of mast cells remain controversial,36-38 there is solid evidence that the Cebpa gene is repressed throughout mast cell development.23,24,34,36 We speculate that the antagonism between GATA2 and C/EBPα may not be involved in the basophil-mast cell fate decision, because both factors were shown to be required for normal basophil development. MITF and/or Ikaros might play a major role in repressing the Cebpa gene at this stage. It is conceivable that GATA2 represses the Cebpa gene in the earlier developmental stage(s) to form a bipotent basophil-mast cell progenitor (BMP) or pre-BMP, via suppression of the monocyte-neutrophil differentiation. This “stepwise replacement” of a negative regulator of the Cebpa gene during mast cell development seems to be a plausible hypothesis that should be examined in further studies. In addition, the molecular mechanism by which the Cebpa gene is repressed in PMCs and other tissue mast cells remains to be elucidated.
The reprogramming of BMMCs was previously reported by Cantor et al in BMMCs transduced with FOG-1, a cofactor for GATA factors.39 They showed that GATA1 functions independently of FOG-1 in mast cells, and FOG-1 downregulation is necessary for mast cell development. Interestingly, the FOG-1-transduced BMMCs had similar morphological features to the ΔCF/ΔCF cells. However, these cells showed much lower cellular viability and a greater incidence of apoptosis compared with the ΔCF/ΔCF cells. Another difference between the FOG-1-transduced and ΔCF/ΔCF BMMCs is their potential to differentiate into erythroid cells. The FOG-1-transduced cells were stimulated by EPO and differentiated into the Ter119-positive erythroid lineage, albeit at a low frequency. In contrast, the expression of EpoR never appeared on the ΔCF/ΔCF cells (data not shown). FOG-1 interacts with both GATA1 and GATA2, both of which are expressed in BMMCs. We previously reported that both factors contribute to the expression of mast cell-specific genes.4 Thus, we consider that the differences might be due to the residual GATA1 activity.
Importantly, our data showed that reduced expression of c-kit and 2 mast cell-specific genes was consistently observed in the ΔCF/ΔCF BMMCs and PMCs. Taken together, these results suggest that GATA2 maintains the mast cell identity by preventing the Cebpa gene expression in a restricted developmental window and plays an essential role in the regulation of mast cell-specific genes throughout development of this cell lineage.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Sally A. Camper (University of Michigan) for providing the Gata2flox/flox mice, Drs Toshiya Inaba and Akinori Kanai for the help performing the A-seq analysis, and Dr Toshio Kitamura for providing the PlatE and pMXs IG vector for the retroviral transduction. The authors also thank Takuya Matsumoto for technical assistance.
This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology (K.O.), a Grant-in-Aid for Scientific Research (C) (K.O.), and a Grant-in-Aid for Young Scientists (B) (S.O.) from the Japan Society for the Promotion of Science.
Authorship
Contribution: S.O. and K.O. designed the study; S.O. performed most of the experiments; K.O., Y.I., K.K., Y.N., M.I., and N.T. performed several experiments; S.O. and K.O. analyzed and interpreted the data and wrote the manuscript; and O.O., T.M., and M.Y. helped with the data interpretation and the writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kinuko Ohneda, Department of Pharmacy, Faculty of Pharmacy, Takasaki University of Health and Welfare, 60 Nakaorui-machi, Takasaki-shi, Gunma 370-0033, Japan; e-mail: kohneda@takasaki-u.ac.jp.

![Figure 1. The deletion of GATA2 CF results in a loss of mast cell identity in mouse bone marrow-derived mast cells. (A) (Left) Representative FACS data for BMMCs prepared from Gata2flox/flox::Rosa26CreERT2 mice after 0, 3, 6, and 9 days of 4-OHT treatment (D0, D3, D6, and D9, respectively). (Right) Average percentages and SD of the DP cells within the gates examined on days 0, 3, 5, 7, and 9 of the 4-OHT treatment (D0, D3, D5, D7, and D9, respectively). N = 9. (B) Representative bright field (BF) phase contrast photomicrographs (left; original magnification, ×200) and cytospin preparations (center and right; original magnification, ×400) of BMMCs prepared from Gata2+/+::Rosa26CreERT2 (CreERT2) and Gata2flox/flox::Rosa26CreERT2(ΔCF/ΔCF) mice treated with 0.5 μM of 4-OHT for 10 days. For cytospin preparations, the cells were stained with Wright-Giemsa stain (center, WG) and toluidine blue (right, TB). (C) The ratio of 4-OHT (+) relative to 4-OHT (−) BMMCs prepared from CreERT2, ΔCF/+, and ΔCF/ΔCF mice. The cells were counted on days 3 and 6 of the 4-OHT treatment (D3 and D6, respectively). (D) The percentages of dead cells in the CreERT2 and ΔCF/ΔCF BMMCs cultured in the absence (−) or presence (+) of 4-OHT for 3 and 6 days (D3 and D6, respectively). In C and D, no cells survived in the absence of IL-3 and SCF [cytokine(−)]. (E) (Left) Representative FACS data for the 5-bromo-2'-deoxyuridine and 7-amino-actinomycin D expression of ΔCF/ΔCF BMMCs on days 0, 4, and 8 of 4-OHT treatment (D0, D4, and D8, respectively). The lower right panel shows the gates displaying the cell cycle populations in the subG0/G1 (Sub), G0/G1, S, and G2/M phases. (Right) Average percentages and SD of the gated cells. *P < .05, **P < 0.01. N = 4. (F) The results of a FACS analysis of the presorted c-Kit/FcεRIα DP BMMCs from Gata2flox/flox::Rosa26CreERT2 mice after 0, 3, 6, and 9 days of 4-OHT treatment (D0, D3, D6, and D9, respectively). The average percentages and SD of the gated cells are indicated. N = 4.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/21/10.1182_blood-2014-11-612465/4/m_3306f1.jpeg?Expires=1764965465&Signature=UYuY3vApZQP63iOAjW-h8qCW83uSCNHYT4aWw7tWJirhYIuvkgroC8yAUALnURcmvHIlXrqFqelhtOpS5C45ZqZdPlRukTGOt9vMScMx~fZzr-EEo1ehsMfqeBn27wJ-KilB~0Rhalo7JVBvmxMXaxdfG~tDtUd-oQJrASgO~LNN0eQizdBI3vfe7ec35snnvG3Bp9AX05x07BBOZrRuf0YZ~fl6z6xlit4Yz0zLYrlcItRlbR~8YjtzA5uU1PD50Jjhnyf~1E03aB3AqDMjIkda58yB48t3sqsd12D23mc5QikaZn-g5cfbzG~tr4HdIa3NDts5xUhZCvuN7J-y7A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. No upregulation of Cebpa is observed in ΔCF/ΔCF peritoneal mast cells. (A) Representative FACS data for markers of mast cells (c-Kit and FcεRIα) and basophils (CD11b and CD49b) on ΔCF/ΔCF PMCs. PMCs were cultured ex vivo for 14 days and then were cultured in the absence (−) or presence (+) of 4-OHT for an additional 9 days. The average percentages and SD of the DP cells within the gates are shown. N = 4. (B) The results of the Q-PCR analyses of the indicated genes in PMCs and BMMCs prepared from wild-type mice (WT) and ΔCF/ΔCF PMCs cultured in the absence (−) or presence (+) of 4-OHT. *P < .05; **P < .01 [compared with data from 4-OHT(−) ΔCF/ΔCF PMCs]. N = 4. (C-D) Schematic summaries of the present study. (C) Summary of the reprogramming activity of ΔCF/ΔCF BMMCs. (D) Summary of the phenotypes of the genetically manipulated BMMCs and PMCs used in this study.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/21/10.1182_blood-2014-11-612465/4/m_3306f7.jpeg?Expires=1764965465&Signature=PIa10M42MskEI1I6cjkkhUtvAc~Ve1wt4bCPkiKjqp-2H28KJ7b3Z~OLvWkaummj1yWqzWXP~s73GmOe9kJ8f9l-vnoCqKJxp82FTIkIGLUhgrf0Qk8pkJ1YCDtFLqTXAMJeFEMAwpmcwYWYaKH~fTR~XUtAX-ueFHiZ6nLs8P78LpFqOVavLEoh8ytKh60qvI9JWTttU5ZuerJnO~JfnsE1quYp-pSA5-J9MLdFTjs3TDzURxv~rVqy3icjYIVO8ZRm0MT8NiQU68wMpDSJcQcX0WobgmvFaYZvAmjcQ81Z9h1gb0P8pKh1s0vGGf1zHD-DPh3gG2XF1zCcNZ3uwg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal