Key Points
CHD4 depletion sensitizes AML cells but not normal CD34+ progenitors to genotoxic agents by relaxing chromatin and impairing DSB repair.
CHD4 depletion modulates expression of AML cell genes that regulate tumor formation in vivo and colony formation in vitro.
Abstract
Chromodomain helicase DNA-binding protein 4 (CHD4) is an ATPase that alters the phasing of nucleosomes on DNA and has recently been implicated in DNA double-stranded break (DSB) repair. Here, we show that depletion of CHD4 in acute myeloid leukemia (AML) blasts induces a global relaxation of chromatin that renders cells more susceptible to DSB formation, while concurrently impeding their repair. Furthermore, CHD4 depletion renders AML blasts more sensitive both in vitro and in vivo to genotoxic agents used in clinical therapy: daunorubicin (DNR) and cytarabine (ara-C). Sensitization to DNR and ara-C is mediated in part by activation of the ataxia-telangiectasia mutated pathway, which is preliminarily activated by a Tip60-dependent mechanism in response to chromatin relaxation and further activated by genotoxic agent–induced DSBs. This sensitization preferentially affects AML cells, as CHD4 depletion in normal CD34+ hematopoietic progenitors does not increase their susceptibility to DNR or ara-C. Unexpectedly, we found that CHD4 is necessary for maintaining the tumor-forming behavior of AML cells, as CHD4 depletion severely restricted the ability of AML cells to form xenografts in mice and colonies in soft agar. Taken together, these results provide evidence for CHD4 as a novel therapeutic target whose inhibition has the potential to enhance the effectiveness of genotoxic agents used in AML therapy.
Introduction
Acute myeloid leukemia (AML) is a malignancy that arises from the impaired differentiation of hematopoietic progenitors, resulting in the accumulation of immature myeloid blasts in the bone marrow. Despite advances in our understanding and management of AML, the overall 5-year survival is only 24% due to the low remission and high relapse rates in older patients and those with complex tumor genotypes.1 Current AML management begins with induction chemotherapy, generally consisting of an anthracycline, such as daunorubicin (DNR), supplemented with cytarabine (ara-C). Combined DNR/ara-C regimens achieve overall complete remission (CR) rates ranging from 53% to 58%,2 although recent studies suggest that increasing the DNR dose may yield tangible benefits to the CR rates and survival of a subset of patients.3,4 Regimens containing various combinations of anthracyclines and high-dose ara-C also form the foundation of salvage therapy for patients with relapsed disease.5,6
The anthracycline DNR is a topoisomerase inhibitor that induces DNA double-stranded breaks (DSBs), which are highly cytotoxic.7 Similarly, the nucleoside analog ara-C also induces DNA damage, including DSBs, during DNA synthesis through inhibition of DNA polymerase and incorporation into DNA.8,9 Both normal and leukemic cells can evade cell death following chemotherapy-induced DSBs by repairing damage through numerous repair mechanisms. However, malignant cells tend to be more susceptible to DSB insults due to their rapid proliferation, deregulated cell-cycle checkpoints, and inactive DNA repair machinery.10 Recently, the chromatin structure surrounding a DSB has emerged as a key determinant of the kinetics and mechanism of repair. Thus, chromatin-remodeling enzymes that regulate chromatin architecture have been identified as key mediators of the DSB repair process, as they are needed to relax the chromatin structure surrounding DSBs for efficient repair to occur.11-14
Chromodomain helicase DNA-binding protein 4 (CHD4) is a widely conserved member of the sucrose nonfermenting 2 superfamily of chromatin-remodeling ATPases that is capable of altering the phasing of nucleosomes on DNA.15-18 This ATPase is a core subunit of the corepressor nucleosome remodeling and deacetylase (NuRD) complex that has been shown to play a significant role in DNA methylation-dependent transcriptional repression, particularly the repression of hypermethylated tumor suppressor genes in cancer.19-21 Additionally, recent studies found that CHD4 is rapidly recruited to sites of DSBs,22,23 where it facilitates an E3 ubiquitin ligase RNF8-dependent relaxation of the surrounding chromatin to promote the recruitment of other repair machinery, such as RNF168 and BRCA1.24
In this study, we investigate the dual functionality of CHD4 in the context of AML. We show that CHD4 is necessary for the efficient repair of DSBs within AML cells, and that AML cells partially depleted of CHD4 are more susceptible to clinically used DNA-damaging agents, such as DNR and ara-C, both in vitro and in vivo. Additionally, we demonstrate that the depletion of CHD4 in AML cells reduces their potential to form mouse xenografts and markedly inhibits their ability to generate colonies. We show that both of these phenotypes are consistent with gene expression alterations resulting from CHD4 depletion. Importantly, all of these events occur preferentially in AML cells, as similar depletion of CHD4 in normal CD34+ hematopoietic progenitor cells does not result in similar phenotypes. Taken together, the present findings highlight for the first time the therapeutic potential of targeting CHD4 in AML.
Methods
Cells
Human AML U937 and MV4-11 cells were described previously.25 OCI-AML3 cells were purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ; Braunschweig, Germany). Cells were cultured in RPMI 1640 (Gibco) supplemented with 10% fetal bovine serum (FBS; Atlas) and 2% penicillin/streptomycin. Cell numbers were measured by a Cellometer Auto T4 (Nexcelom Bioscience).
Isolation of primary cells
This study was approved by the Virginia Commonwealth University (VCU) Investigational Review Board and samples were collected with patient consent. Bone marrow was collected from AML patients with high disease load (>90% blasts in the marrow) and mononuclear cells were isolated by Ficoll-Hypaque gradient separation as described previously.25 Normal CD34+ hematopoietic progenitor cells were isolated from deidentified apheresis units discarded by the VCU Bone Marrow Transplant Unit as described previously.26 All primary cells were cultured in StemSpan serum-free expansion medium with 1× CC100 cytokine mix (StemCell Technologies) and 2% penicillin/streptomycin.
Short hairpin RNA (shRNA) constructs were created as described previously.26 Briefly, CHD4 target sequences (shCHD4-1: CGGTGAGATCATCCTGTGTGATA; shCHD4-2: GGACCTGAATGATGAGAAACAGA) and the Tip60 target sequence (CCTCAATCTCATCAACTACTA)27 were cloned into a green fluorescent protein (GFP)-expressing pRRL.H1.shRNA vector and packaged into a lentivirus through calcium phosphate transfections in 293T cells.
Immunoblotting was performed as described previously.28,29 Blots were imaged and quantitated using a Li-Cor Odyssey Fc Imager. Primary antibodies for the antigens studied were obtained from several sources: H3K9ac, H3, H4K8ac, H4, cleaved-caspase 3, phosphorylated ataxia-telangiectasia mutated (p-ATM; S1981), and ATM, all from Cell Signaling; CHD4 from Millipore; poly(ADP-ribose) polymerase (PARP) from Biomol Research Laboratories; E2F1 from Santa Cruz Biotechnology; and α-tubulin from Calbiochem.
Confocal microscopy was performed on a Zeiss 720 Meta microscope as previously described.30 Primary antibodies used were against H3K9me3 and γ-H2A.X (Millipore). Images were quantified using the Velocity Image Analysis software (PerkinElmer).
Cell sensitivity to DNR and ara-C
Cells were incubated with DNR (Sigma-Aldrich) or ara-C (Sigma-Aldrich) for 24 hours and a 7-aminoactinomycin D (7AAD; Sigma-Aldrich) stain read by a FACSCanto Cell Analyzer (BD) was used to determine cell viability.
In vivo studies were approved by the VCU Institutional Animal Care and Use Committee. Female NOD scid γ mice (NSG; The Jackson Laboratory) were engrafted IV via the tail vein with 5 × 106 luciferase-expressing U937 cells. Tumor progression was monitored by the total radiance generated within each mouse upon subcutaneous administration of luciferin as measured by the IVIS 200 imaging system (Xenogen Corporation). Once tumor engraftment was confirmed, the mice were treated with a previously described model of the human “7 + 3” induction therapy.33,34 Briefly, this regimen consisted of 3 consecutive days of Doxorubicin (100 mg/kg; Selleck Chemicals) and ara-C (33 mg/kg; Selleck Chemicals) given IV by tail-vein injection, followed by 2 additional days of ara-C (33 mg/kg) given by intraperitoneal injection.
Colony-forming assays were performed as previously described.28 For AML cell lines, 5000 cells were suspended in 1 mL of 1× RPMI supplemented with 0.35% agarose (Difco), 10% FBS, and 2% penicillin/streptomycin and plated into a well of a 12-well plate. Cells were allowed to grow for 14 days and colonies were identified as having >50 cells. For CD34+ cells, the Methocult H4434 Classic kit (StemCell Technologies) was used.
Microarray analysis was performed using HG-U133Ax2 GeneChips (Affymetrix). Quality of the hybridization was assessed by examining the average background, scaling factor, the percentage of probe sets called present by the detection call algorithm, and the 3′:5′ ratio for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ACTIN. Each GeneChip was independently normalized using quantile normalization and the robust multiarray average method was applied to probe-set expression summaries.35 For each probe set, a moderated t test was used to make comparisons using the limma Bioconductor package36,37 in the R programming environment.38 The P values were used in estimating the false discovery rate (FDR) using the Benjamini and Hochberg method39 and probe sets having an FDR < 0.05 were considered significant. Pathway analysis of the significant probe sets was performed using the Ingenuity Pathway Analysis software (Qiagen).
Statistical analysis
Results are shown as means + standard deviation (SD) unless otherwise stated. P values were calculated by the Student t test.
Results
CHD4 is necessary for maintaining heterochromatin in AML blasts
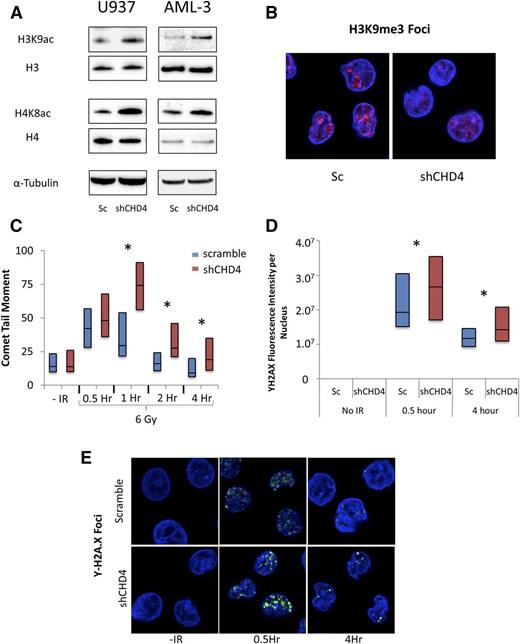
As CHD4 is a chromatin-remodeling ATPase,15-18 we investigated the effect of CHD4 depletion on the overall chromatin structure of AML cells. Integration of a CHD4-targeting shRNA (shCHD4-1) into the AML cell lines U937, MV4-11, and AML-3 by a lentiviral vector resulted in the robust depletion of the CHD4 protein (supplemental Figure 1A, see supplemental Data available at the Blood Web site). When compared with control cells in which a scrambled, nontargeting shRNA was integrated, cells depleted of CHD4 were found to have a global increase in the euchromatin-associated acetylation of histone H3 lysine 9 and histone H4 lysine 8 (Figure 1A; supplemental Table 1) and disruption of heterochromatin-associated trimethyl-histone H3 lysine 9 foci (Figure 1B). These results indicate that inhibition of CHD4 function induces a global relaxation of the chromatin structure in AML blasts.
CHD4 is necessary for the maintenance of heterochromatin and the efficient repair of DNA DSBs in AML blasts. AML cell lines were infected by lentivirus to integrate either a nontargeting, scrambled (Sc) shRNA or a CHD4-targeting shRNA. (A) Depletion of CHD4 leads to a global increase in euchromatin-associated histone H3K9 and H4K8 acetylation. (B) Depletion of CHD4 also results in a disruption of heterochromatin-associated histone H3K9me3 foci in U937 cells as shown by staining with anti-H3K9me3 antibody. (C) U937 cells were exposed to 6 Gy of radiation to induce DNA DSBs. DSB formation and repair was monitored over a 4-hour time course using a neutral comet assay. CHD4-depleted cells displayed significantly more evidence of DSBs and were delayed in their repair (n > 50 comets). (D) The results of the comet assay were confirmed by staining for Υ-H2A.X foci and calculating the fluorescent intensity per nucleus (n > 60 cells per time point). (E) Representative images of the Υ-H2A.X stain. *P < .05.
CHD4 is necessary for the maintenance of heterochromatin and the efficient repair of DNA DSBs in AML blasts. AML cell lines were infected by lentivirus to integrate either a nontargeting, scrambled (Sc) shRNA or a CHD4-targeting shRNA. (A) Depletion of CHD4 leads to a global increase in euchromatin-associated histone H3K9 and H4K8 acetylation. (B) Depletion of CHD4 also results in a disruption of heterochromatin-associated histone H3K9me3 foci in U937 cells as shown by staining with anti-H3K9me3 antibody. (C) U937 cells were exposed to 6 Gy of radiation to induce DNA DSBs. DSB formation and repair was monitored over a 4-hour time course using a neutral comet assay. CHD4-depleted cells displayed significantly more evidence of DSBs and were delayed in their repair (n > 50 comets). (D) The results of the comet assay were confirmed by staining for Υ-H2A.X foci and calculating the fluorescent intensity per nucleus (n > 60 cells per time point). (E) Representative images of the Υ-H2A.X stain. *P < .05.
Depletion of CHD4 renders AML blasts more sensitive to DSB damage and impairs the repair of DSBs
Because chromatin-remodeling enzymes have been found to play key roles in mediating the repair of DSBs,11,14 we investigated whether CHD4 is involved in DSB repair in AML blasts. We subjected U937 cells to 6 Gy of radiation and detected DSBs using a neutral comet assay over a 4-hour time course (Figure 1C). CHD4-depleted cells demonstrated significantly more evidence of DSBs 1 hour after radiation exposure, suggesting that inhibition of CHD4 rendered the cells more susceptible to the formation of DSBs. The tail moment of control cells returned to baseline after just 2 hours, indicating that cells expressing wild-type levels of CHD4 are capable of rapidly repairing radiation-induced DSBs to levels below the detection limits of the assay. However, the tail moment of the CHD4-depleted cells remained elevated throughout the 4-hour course of the experiment, indicating that CHD4 is required for the efficient repair of DSBs in AML cells.
To confirm our observations from the neutral comet assay, we performed immunostaining for γH2A.X foci, a histone marker of DSBs, over the 4-hour time course (Figure 1D-E). Cells depleted of CHD4 contained a significant increase in γH2A.X foci at 0.5 hours postradiation exposure and the number of foci remained elevated above that in control cells over the 4-hour time course. Similar results were obtained for MV4-11 and AML-3 (supplemental Figure 2).
We then confirmed that the DSBs observed over the 4-hour time course were not the result of apoptosis, as neither PARP nor caspase-3 cleavage occurred throughout the 4-hour time course (supplemental Figure 3). Thus, in AML cells, inhibition of CHD4 impedes the repair of DSBs and increases susceptibility to their formation.
Inhibition of CHD4 renders AML blasts more sensitive to DNR and ara-C in vitro
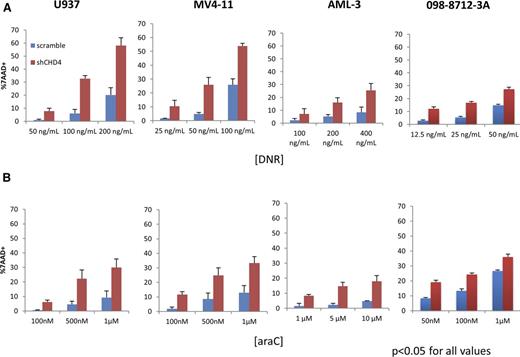
Having determined that CHD4 affects the formation and repair of DSB in AML cells, we hypothesized that AML cells depleted of CHD4 would be more sensitive to the DSB-inducing agents DNR and ara-C. To test this hypothesis, we incubated the AML cells for 24 hours with clinically relevant concentrations of either DNR (Figure 2A) or ara-C (Figure 2B) and the assessed their viability with 7AAD staining. At all DNR and ara-C concentrations tested, significantly more cell death was observed in the AML cells depleted of CHD4, with some concentrations resulting in as much as a fivefold increase in cell death. Similar results were obtained when we depleted CHD4 in a primary AML sample (098-8712-3A). To confirm that the increase in sensitivity to DNR and ara-C was not the result of an off-target effect of the shRNA construct used, a second CHD4-targeted shRNA construct (shCHD4-2; supplemental Figure 1A-B) was tested and induced a similar increase in sensitivity to DNR (supplemental Figure 4). Thus, knockdown of CHD4 with 2 independent shRNA constructs enhanced the in vitro sensitivity of AML cells to DNR and ara-C.
Inhibition of CHD4 renders AML blasts more sensitive to DNR and ara-C in vitro. U937, MV4-11, and AML-3 human AML cell lines and a primary AML sample were incubated with various concentrations of (A) DNR or (B) ara-C for 24 hours in vitro. Cell viability was assayed using a 7AAD stain and quantified by flow cytometry. For all AML cells and concentrations of DNR/ara-C tested, AML cells depleted of CHD4 displayed significantly more cell death compared with controls. P < .05 for every data point tested; n = 3 biological replicates for each data point.
Inhibition of CHD4 renders AML blasts more sensitive to DNR and ara-C in vitro. U937, MV4-11, and AML-3 human AML cell lines and a primary AML sample were incubated with various concentrations of (A) DNR or (B) ara-C for 24 hours in vitro. Cell viability was assayed using a 7AAD stain and quantified by flow cytometry. For all AML cells and concentrations of DNR/ara-C tested, AML cells depleted of CHD4 displayed significantly more cell death compared with controls. P < .05 for every data point tested; n = 3 biological replicates for each data point.
As DNR is a fluorescent-intercalating agent40 and CHD4 depletion induces a global relaxation of chromatin in AML blasts, we tested whether CHD4-depleted cells were more susceptible to DNR intercalation. Incubation of U937 cells for 2 hours with 200 ng/mL DNR caused significantly more DNR fluorescent signal inside the nuclei of CHD4-depleted cells (supplemental Figure 5). This finding is consistent with increased DNR intercalation within CHD4-depleted cells.
Inhibition of CHD4 does not sensitize normal CD34+ progenitor cells to DNR and ara-C
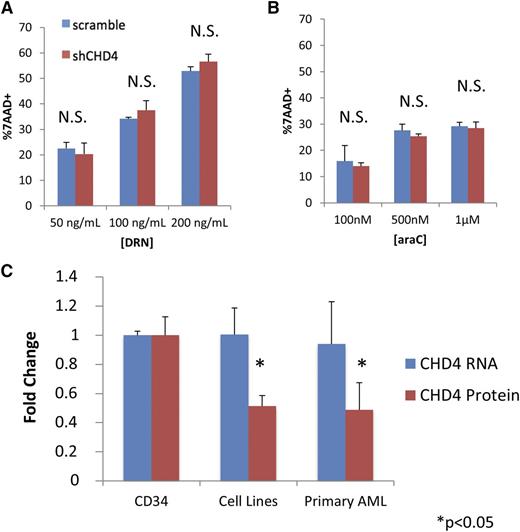
We next investigated whether the increased sensitivity to DNR and ara-C induced by CHD4 inhibition occurs preferentially in AML blasts. We depleted CHD4 in CD34+ hematopoietic progenitor cells isolated from the mobilized blood of 3 normal donors (supplemental Figure 1E). Following incubation for 24 hours with DNR and ara-C, CD34+ cells did not display a significant increase in sensitivity to DNR (Figure 3A) or ara-C (Figure 3B).
Inhibition of CHD4 does not sensitize normal CD34+ progenitor cells to DNR and ara-C. CD34+ hematopoietic progenitors were isolated from the 3 normal donors. Depletion of CHD4 did not significantly increase the sensitivity of the CD34+ cells to either (A) DNR or (B) ara-C. (C) The CD34+ cells were found to contain significantly more endogenous CHD4 protein than the AML cell lines (U937, MV4-11, AML-3) and primary AML blasts isolated from the bone marrow of 3 patients. Interestingly, there was no difference in CHD4 mRNA between the CD34+ and AML cells. *P < .05. N.S., not statistically significant.
Inhibition of CHD4 does not sensitize normal CD34+ progenitor cells to DNR and ara-C. CD34+ hematopoietic progenitors were isolated from the 3 normal donors. Depletion of CHD4 did not significantly increase the sensitivity of the CD34+ cells to either (A) DNR or (B) ara-C. (C) The CD34+ cells were found to contain significantly more endogenous CHD4 protein than the AML cell lines (U937, MV4-11, AML-3) and primary AML blasts isolated from the bone marrow of 3 patients. Interestingly, there was no difference in CHD4 mRNA between the CD34+ and AML cells. *P < .05. N.S., not statistically significant.
To gain insight into how CHD4 depletion might preferentially sensitize leukemic cells, we compared the total endogenous CHD4 protein levels in CD34+ cells from the 3 normal donors to that in 3 AML cell lines (U937, MV4-11, AML-3) and blasts isolated from the bone marrow of 3 different AML patients whose marrow contained >90% blasts. AML cell lines and primary AML blasts contained on average ∼50% less CHD4 total protein than CD34+ cells (Figure 3C; supplemental Figure 1E). Surprisingly, AML cell lines and primary AML samples did not have significantly different amounts of CHD4 messenger RNA (mRNA) compared with the normal CD34+ progenitors, suggesting that posttranscriptional alterations could account for the discrepancy in the observed amount of CHD4 protein.
Inhibition of CHD4 sensitizes AML blasts to DNR and ara-C in a xenograft model
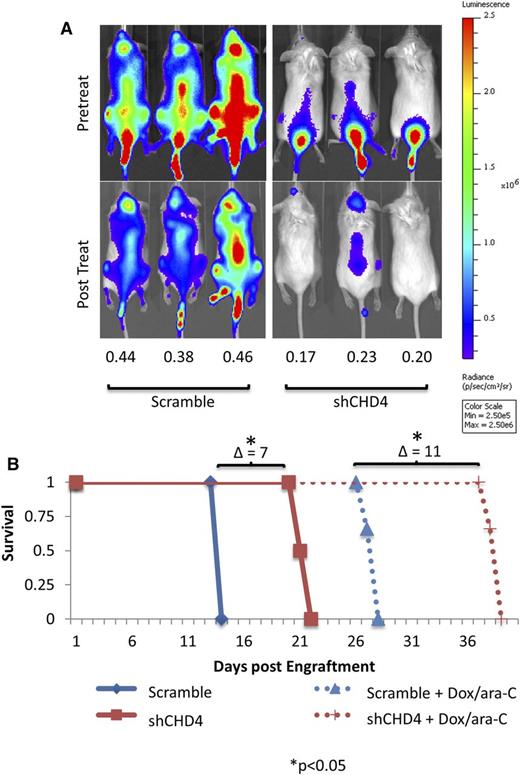
Having demonstrated that depletion of CHD4 preferentially increases the sensitivity of AML cells to DNR and ara-C in vitro, we tested the effect of CHD4 depletion in AML cells in vivo. To this end, we used a previously described mouse xenograft model of the human “7 + 3” induction regimen33,34 that consists of 5 continuous days of ara-C treatment and doxorubicin coadministered on days 1 through 3. In this model, the anthracycline doxorubicin is substituted for the anthracycline DNR used in humans due to the systemic toxicity of the latter in mice.33 Luciferase-expressing U937 cells were systemically engrafted by tail-vein injection into NSG mice (n = 3 mice per condition) and disease progression was monitored by total radiance (measured in photons/second/centimeter2/steradian) generated within each mouse upon subcutaneous administration of luciferin.
The treatment regimen was initiated once a systemic xenograft was confirmed and mice were reimaged 7 days later, a time we had previously observed to yield the maximum drug-induced antitumor effect. Posttreatment, mice engrafted with control cells displayed, on average, 43% of their total pretreatment radiance. However, mice engrafted with CHD4-depleted cells displayed only 20% of their pretreatment radiance (Figure 4A), indicating that AML blasts depleted of CHD4 are indeed more sensitive to DNR and ara-C treatment in vivo.
Inhibition of CHD4 sensitizes AML blasts to DNR and ara-C in a xenograft model. Equal numbers of luciferase-expressing Scramble and shCHD4 U937 cells were engrafted into NSG mice by tail-vein injection and tumor burden was noninvasively monitored by measuring the total radiance generated upon subcutaneous administration of luciferin. (A) Once a tumor was well established (as seen in the pretreatment panels), the mice were treated with 5 continuous days of ara-C, with concurrent doxorubicin on days 1 through 3. Posttreatment, mice engrafted with control cells were found to possess 43% of their pretreatment radiance, whereas mice engrafted with CHD4-depleted cells possessed 20% (P < .001). (B) The mice were followed for survival. CHD4 depletion alone resulted in a 7-day improvement in mouse survival. Treatment with Doxorubicin/ara-C improved the survival of the mice engrafted with CHD4-depleted cells by an additional 4 days. *P < .05.
Inhibition of CHD4 sensitizes AML blasts to DNR and ara-C in a xenograft model. Equal numbers of luciferase-expressing Scramble and shCHD4 U937 cells were engrafted into NSG mice by tail-vein injection and tumor burden was noninvasively monitored by measuring the total radiance generated upon subcutaneous administration of luciferin. (A) Once a tumor was well established (as seen in the pretreatment panels), the mice were treated with 5 continuous days of ara-C, with concurrent doxorubicin on days 1 through 3. Posttreatment, mice engrafted with control cells were found to possess 43% of their pretreatment radiance, whereas mice engrafted with CHD4-depleted cells possessed 20% (P < .001). (B) The mice were followed for survival. CHD4 depletion alone resulted in a 7-day improvement in mouse survival. Treatment with Doxorubicin/ara-C improved the survival of the mice engrafted with CHD4-depleted cells by an additional 4 days. *P < .05.
Mice were then followed for survival, until a humane end point was reached (Figure 4B). Untreated mice engrafted with CHD4-depleted cells survived significantly longer than those engrafted with control cells. The survival of the mice engrafted with CHD4-depleted cells further improved with the addition of the drug regimen. Once the mice reached their humane end points, bone marrow was collected and analyzed. In the marrow of mice that received the shCHD4-depleted cells, the AML cells retained ∼75% knockdown of CHD4 mRNA compared with cells from the bone marrow of mice containing the control shRNA (data not shown).
Inhibition of CHD4 improves survival in mice
Despite injection with equivalent numbers of viable leukemic cells at the same time, mice engrafted with CHD4-depleted cells harbored significantly lower tumor burden than mice engrafted with control cells when therapy was initiated. CHD4 depletion did not significantly alter the in vitro (supplemental Figure 6) or in vivo (supplemental Figure 7) proliferation rate, nor did it alter the cell-cycle distribution of the tested AML cell lines (supplemental Figure 8). Therefore, proliferative alterations are unlikely to account for the discrepancy in the tumor burden between mice engrafted with control vs CHD4-depleted cells.
An alternative explanation is that CHD4 depletion reduces the tumor-forming capacity of the leukemic cells. We performed soft agar colony-forming assays, as leukemic cells capable of forming colonies in soft agar have been shown to possess the ability to initiate disease in vivo.41 Indeed, CHD4 depletion sharply reduced the formation of AML colonies in soft agar (Figure 5A-B; supplemental Figure 9), suggesting that CHD4 is required to maintain the full tumor-forming ability of these cells.
CHD4 depletion dramatically decreases the tumor-forming behavior of AML cells and modulates expression of genes associated with tumor colony formation. CHD4-depleted cells were inhibited in their ability to engraft into the NSG mice. CHD4 depletion severely reduced the ability of U937 and MV4-11 cell lines to form colonies in soft agar. (A) Representative images (20×) of colonies. (B) Quantitation of AML colonies (*P < .05, n = 3). (C) Microarray arrays were performed to determine the gene expression alterations associated with CHD4 depletion. Ingenuity Pathway Analysis software was used to interpret the data and predicted a significant decrease in the colony-forming potential in both U937 and MV4-11 (activation Z score of −2.044 and −2.002, respectively). (D) Depletion of CHD4 in CD34+ hematopoietic progenitors isolated from 3 normal donors did not result in a significant decrease in colony formation.
CHD4 depletion dramatically decreases the tumor-forming behavior of AML cells and modulates expression of genes associated with tumor colony formation. CHD4-depleted cells were inhibited in their ability to engraft into the NSG mice. CHD4 depletion severely reduced the ability of U937 and MV4-11 cell lines to form colonies in soft agar. (A) Representative images (20×) of colonies. (B) Quantitation of AML colonies (*P < .05, n = 3). (C) Microarray arrays were performed to determine the gene expression alterations associated with CHD4 depletion. Ingenuity Pathway Analysis software was used to interpret the data and predicted a significant decrease in the colony-forming potential in both U937 and MV4-11 (activation Z score of −2.044 and −2.002, respectively). (D) Depletion of CHD4 in CD34+ hematopoietic progenitors isolated from 3 normal donors did not result in a significant decrease in colony formation.
CHD4 depletion alters expression of genes involved in tumor formation
To gain insight into the molecular basis for this loss of colony-forming potential, we performed microarrays to investigate transcriptional alterations that result from the depletion of CHD4. Probe sets were considered to be differentially expressed if the FDR was <0.05, which resulted in 1320 and 675 differentially expressed genes in U937 and MV4-11, respectively. We then used a downstream effects analysis in the Ingenuity Pathway Analysis (IPA) software to interpret the biological implications of gene expression alterations (Figure 5C). According to this analysis, CHD4 depletion resulted in a net decrease in the expression of genes previously reported to increase the colony-forming potential of tumor cells and concomitantly induced the expression of genes previously reported to decrease the colony-forming potential of tumor cells. Taken together, the IPA analysis predicted a significant decrease in the colony formation potential of both U937 and MV4-11 cells.
We then used quantitative polymerase chain reaction to validate the expression alterations of several genes whose differential expression has previously been shown to significantly alter the colony formation of AML and hematopoietic stem cells, including Myc and PCGF2.42,-44 Consistent with the microarray data, Myc was found to be downregulated and PCGF2 was found to be upregulated in the CHD4-depleted AML cells (supplemental Figure 10). An additional 7 genes were also validated in CHD4-depleted AML cells (supplemental Figure 11). In conjunction with the IPA analysis, these results indicate that transcriptional alterations induced by the depletion of CHD4 can at least partially explain the decrease in colony-forming potential in AML cells.
In contrast to the findings in AML cells, depletion of CHD4 did not significantly alter the colony-forming potential of CD34+ cells isolated from normal donors (Figure 5D; supplemental Figure 12). Consistent with this observation, the expression of neither Myc nor PCGF2 was significantly altered in the CHD4-depleted CD34+ cells (supplemental Figure 10).
Inhibition of CHD4 activates the ATM signaling pathway to induce apoptosis
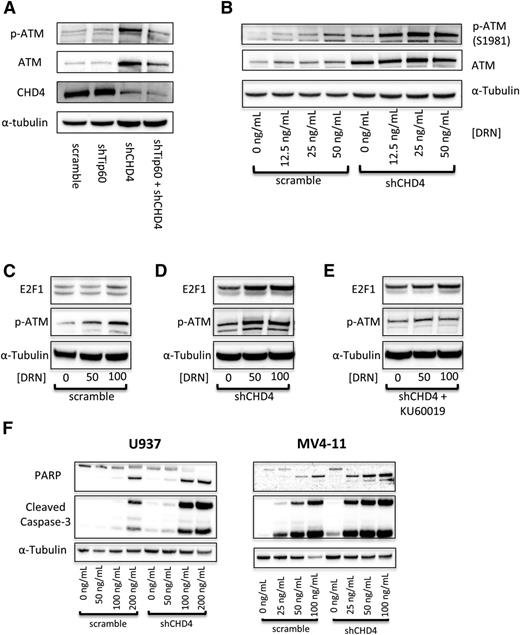
As AML cells depleted of CHD4 are more sensitive to DSB-inducing agents, we investigated the role of the ATM pathway. The serine/threonine kinase ATM is a master regulator of the DSB repair response and is classically activated by autophosphorylation of serine 1981 in response to double-stranded DNA breaks.10,45 Recently, the protein acetyl-transferase Tip60 was found to activate ATM in response to chromatin relaxation.46 Because depletion of CHD4 led to a global relaxation of the chromatin structure of AML cells, we hypothesized that depletion of CHD4 would induce a Tip60-dependent activation of ATM. Accordingly, a 10-fold increase in the phosphorylation of S1981 on ATM in U937 cells was observed upon the depletion of CHD4. However, the phosphorylation of ATM remained at control levels upon concomitant depletion of Tip60 in CHD4-depleted cells (Figure 6A; supplemental Table 2). This observation indicates that inhibition of CHD4 does induce a Tip60-dependent activation of ATM.
Inhibition of CHD4 activates the ATM signaling pathway to induce increased apoptosis. (A) Depletion of CHD4 in U937 cells induces a significant activation of ATM, which is blocked by the concurrent depletion of Tip60. (B) Upon addition of DNR, ATM is activated in a concentration-dependent manner, with significantly more activation in the CHD4-depleted cells. (C) Once active, ATM acts on its downstream targets, including stabilizing the proapoptotic transcription factor E2F1. E2F1 is stabilized in a DNR concentration-dependent manner, (D) with significantly more being stabilized in the CHD4-depleted cells. (E) This stabilization is diminished upon the addition of the ATM inhibitor KU60019. (F) Ultimately, in response to the DNR, CHD4-depleted AML cells display elevated markers of apoptosis, including PARP and Caspase-3 cleavage.
Inhibition of CHD4 activates the ATM signaling pathway to induce increased apoptosis. (A) Depletion of CHD4 in U937 cells induces a significant activation of ATM, which is blocked by the concurrent depletion of Tip60. (B) Upon addition of DNR, ATM is activated in a concentration-dependent manner, with significantly more activation in the CHD4-depleted cells. (C) Once active, ATM acts on its downstream targets, including stabilizing the proapoptotic transcription factor E2F1. E2F1 is stabilized in a DNR concentration-dependent manner, (D) with significantly more being stabilized in the CHD4-depleted cells. (E) This stabilization is diminished upon the addition of the ATM inhibitor KU60019. (F) Ultimately, in response to the DNR, CHD4-depleted AML cells display elevated markers of apoptosis, including PARP and Caspase-3 cleavage.
We next sought to investigate the ATM pathway in AML cells in response to DNR. As expected, ATM is autophosphorylated in a dose-dependent manner in both control and shCHD4 cells. However, a significant increase in the amount of p-ATM was detected at each DNR concentration tested in CHD4-depleted cells (Figure 6B). Interestingly, depletion of CHD4 also increased the level of total ATM by threefold. Once active, ATM induces numerous downstream phosphorylation events that signal for cell survival by initiating the repair of DSBs or it can trigger apoptosis.10,47 E2F1 is a proapoptotic transcription factor that is stabilized by ATM through phosphorylation of Serine-31.48 E2F1 is stabilized in U937 cells in a DNR dose-dependent manner in control cells (Figure 6C); however, significantly more E2F1 is stabilized in response to DNR in cells partially depleted of CHD4 (Figure 6D). E2F1 did not undergo a DNR dose-dependent stabilization when the ATM inhibitor KU60019 was added, indicating that the observed dose-dependent stabilization requires ATM (Figure 6E). Ultimately, the increased activation of ATM and the subsequently increased downstream proapoptotic signaling in response to DNR in CHD4-depleted AML cells resulted in increased apoptosis compared with control cells (Figure 6F).
Discussion
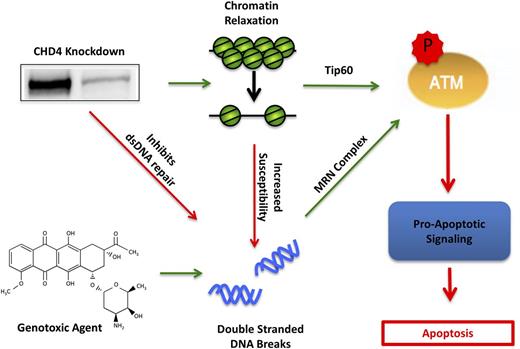
CHD4 and its associated NuRD complex have emerged as key mediators of the double-stranded DNA damage repair pathway11,12 and gene expression, most notably repression of hypermethylated tumor suppressor genes in cancer.19,20 Here, we describe the first study that examines the interplay of CHD4’s dual functionality in AML. We observed that the partial depletion of CHD4 in AML cells induces a global relaxation of the chromatin structure (Figure 1), which is similar to previous reports in Ramos49 and HeLa50 cell lines. CHD4-depleted AML cells were also found to be more susceptible to the formation of DSBs (Figure 1), consistent with evidence that CHD4 protects U2OS osteosarcoma cells from ionizing radiation22,23,51 and that relaxed chromatin is more susceptible to the formation of DSBs than condensed chromatin.52,53 In addition, our results show that the DNA of CHD4-depleted cells were more exposed to intercalation by DNR (supplemental Figure 5), consistent with the greater capacity of anthracyclines to bind nucleosome-free DNA.54 CHD4-depleted AML cells also failed to efficiently repair the resulting genotoxic agent–induced DSBs (Figure 1). Probing ATM pathway components in CHD4-depleted AML cells revealed a Tip60-mediated activation of ATM prior to DNA damage as well as an ATM-dependent stabilization of the proapoptotic transcription factor E2F1 upon induction of DSBs (Figure 6). Taken together, these results indicate that targeting CHD4 could increase the sensitivity of AML cells to genotoxic agents commonly used in therapy through a combination of increased susceptibility to the formation of DSBs and inhibition of their repair (Figure 7).
Model of CHD4 enhancement of genotoxic agent-induced apoptosis through DSB repair inhibition and chromatin relaxation. In AML blasts, depletion of CHD4 relaxes the chromatin, resulting in the activation of ATM through Tip60 and thereby primes the cells for apoptotic signaling. Moreover, the relaxed chromatin is more susceptible to genotoxic agent-induced DSBs. Additionally, CHD4-deficient blasts have impaired DSB repair, resulting in the accumulation of more DSBs and further activation of ATM through the classical Mre11, Rad50, NBs1 (MRN) complex. The net activation of ATM in CHD4-depleted cells increases its downstream proapoptotic signaling cascade, thereby resulting in apoptotic cell death.
Model of CHD4 enhancement of genotoxic agent-induced apoptosis through DSB repair inhibition and chromatin relaxation. In AML blasts, depletion of CHD4 relaxes the chromatin, resulting in the activation of ATM through Tip60 and thereby primes the cells for apoptotic signaling. Moreover, the relaxed chromatin is more susceptible to genotoxic agent-induced DSBs. Additionally, CHD4-deficient blasts have impaired DSB repair, resulting in the accumulation of more DSBs and further activation of ATM through the classical Mre11, Rad50, NBs1 (MRN) complex. The net activation of ATM in CHD4-depleted cells increases its downstream proapoptotic signaling cascade, thereby resulting in apoptotic cell death.
Depletion of CHD4 in 3 AML cell lines and a primary AML patient sample increased the in vitro sensitivity to clinically relevant concentrations of single-agent DNR and ara-C (Figure 2). Importantly, the increased sensitivity to DNR and ara-C occurred preferentially in AML cells, as normal CD34+ hematopoietic progenitors partially depleted of CHD4 did not demonstrate a similar increase in susceptibility (Figure 3). We hypothesize that the preferential sensitivity of AML cells to CHD4 depletion may reflect disparate levels of endogenous CHD4 between normal CD34+ cells and AML cells, as AML cell lines and primary AML cells appear to contain significantly less total endogenous CHD4 protein compared with normal CD34+ progenitor cells (Figure 3C). It remains uncertain whether the diminished levels of CHD4 protein in AML cells reflect the normal differentiation of myeloid lineage cells or are associated in a yet-to-be determined way with the tumorigeneic capacity of AML. In any case, these findings argue that a threshold level of CHD4 may be required for efficient DSB repair, implying that leukemia cells containing less endogenous CHD4 may be more vulnerable to CHD4 inhibition than their normal counterparts.
In a clinically relevant mouse model of induction therapy, NSG mice engrafted with CHD4-depleted U937 cells displayed increased drug sensitivity and survival (Figure 4). An unexpected and important finding was that depletion of CHD4 significantly reduced the tumor burden of the xenografts, which resulted in an additional treatment-independent increase in survival. We also observed that depletion of CHD4 sharply reduces the growth of AML colonies in soft agar, but not the growth of normal donor CD34+ hematopoietic progenitor colonies. This suggests that a minimal level of CHD4 is necessary for the maintenance of tumor-forming behavior of leukemic cells, consistent with evidence that CHD4 and its associated NuRD complex regulate genes implicated in the tumor-initiating state of glioblastoma.55
The role of CHD4 in AML appears to be quite complex, as it exhibits a prooncogenic function through its role in tumor formation, but concurrently displays a tumor suppressor function in its facilitation of DSB repair. We attribute this apparent discrepancy to the diverse roles that CHD4 fulfills within a cell, including regulating gene expression and facilitating DNA repair. This exhibition of both prooncogenic and tumor suppressor properties is shared with other chromatin modifiers such as HDAC156 and EZH257,58 and may reflect a general property of epigenetic regulators. Regardless, the data presented here suggest that CHD4 represents a plausible, new therapeutic target in AML. As combinations of anthracyclines and ara-C form the foundation of both AML induction and salvage therapy, inhibiting CHD4 might enhance the efficacy of current regimens without increasing the required doses of these agents. This would be of particular interest for anthracycline-based regimens, as the maximum lifetime dose of anthracyclines is severely limited by a cumulative, dose-dependent cardiotoxicity,59 although study of CHD4 inhibition in cardiac tissue is needed to corroborate this. Furthermore, the chromatin-relaxation effect of CHD4 inhibition may have additional implications for anthracycline-based therapy, as anthracyclines have been found to displace nucleosomes from transcription start sites of genes located in regions of relaxed chromatin which deregulates the transcriptome and drives apoptosis in AML cells.60 Thus, inhibition of CHD4 may also potentiate DNA damage-independent mechanisms of anthracycline-mediated antileukemic activities. Finally, given the role of CHD4 in maintaining the tumor-forming behavior of AML cells, CHD4 inhibition could have broader implications for disease outcomes in light of the role of tumor-forming behavior in disease relapse.41,61-68
The microarray data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE71865).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Julie Farnsworth for assistance with flow cytometry experiments, Dr Richard Moran, Dr Kristoffer Valerie, and Mary Jeannette Sperlazza for helpful discussions and manuscript critiques, and Amy Jones for assistance in manuscript preparation.
This work was supported by the National Institutes of Health, National Cancer Institute (NIH-NCI) R01 CA 67708 (S.G.) and Leukemia & Lymphoma Society of America grant 6472-15 (S.G.), NIH National Institute of Diabetes and Digestive and Kidney Diseases R01 DK 29902 (G.D.G.), Virginia Commonwealth University (VCU) Massey Cancer Center Support grant from the NCI 5P30 CA016059, and VCU Center for Clinical and Translational Research grant NIH ULITR000058. Services and products in support of the research project were generated by the VCU Massey Cancer Center Flow Cytometry, Biostatistics, and Tissue and Data Acquisition and Analysis Core Shared Resources, supported, in part, with funding from the NIH-NCI Cancer Center Support grant P30 CA016059. Microscopy was performed at the VCU Department of Anatomy and Neurobiology Microscopy Facility and supported, in part, by funding from the NIH National Institute of Neurological Disorders and Stroke Center Core grant 5 P30 NS047463 and, in part, from the NIH-NCI Cancer Center Support grant P30 CA016059.
Authorship
Contribution: J.S., M.R., and G.D.G. designed and planned experiments; J.S., J.B., M.A., E.H., S.Z.W., S.Z.Z., S.P., and C.D. performed experiments; J.S., M.R., K.A., S.G., and G.D.G. analyzed data; and J.S. and G.D.G. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gordon D. Ginder, Massey Cancer Center, Virginia Commonwealth University, 401 College St, Room GRL-135, Richmond, VA 23298; e-mail: gdginder@vcu.edu.