Key Points
EHMT1/2 inhibition increases human γ-globin and HbF expression, as well as mouse embryonic β-globin gene expression.
EHMT1/2 inhibition decreases H3K9Me2 and increases H3K9Ac at the γ-globin gene locus in adult human erythroid cells.
Abstract
Fetal hemoglobin (HbF, α2γ2) induction is a well-validated strategy for sickle cell disease (SCD) treatment. Using a small-molecule screen, we found that UNC0638, a selective inhibitor of EHMT1 and EHMT2 histone methyltransferases, induces γ-globin expression. EHMT1/2 catalyze mono- and dimethylation of lysine 9 on histone 3 (H3K9), raising the possibility that H3K9Me2, a repressive chromatin mark, plays a role in silencing γ-globin expression. In primary human adult erythroid cells, UNC0638 and EHMT1 or EHMT2 short hairpin RNA–mediated knockdown significantly increased γ-globin expression, HbF synthesis, and the percentage of cells expressing HbF. At effective concentrations, UNC0638 did not alter cell morphology, proliferation, or erythroid differentiation of primary human CD34+ hematopoietic stem and progenitor cells in culture ex vivo. In murine erythroleukemia cells, UNC0638 and Ehmt2 CRISPR/Cas9-mediated knockout both led to a marked increase in expression of embryonic β-globin genes Hbb-εy and Hbb-βh1. In primary human adult erythroblasts, chromatin immunoprecipitation followed by sequencing analysis revealed that UNC0638 treatment leads to genome-wide depletion in H3K9Me2 and a concomitant increase in the activating mark H3K9Ac, which was especially pronounced at the γ-globin gene region. In RNA-sequencing analysis of erythroblasts, γ-globin genes were among the most significantly upregulated genes by UNC0638. Further increase in γ-globin expression in primary human adult erythroid cells was achieved by combining EHMT1/2 inhibition with the histone deacetylase inhibitor entinostat or hypomethylating agent decitabine. Our data provide genetic and pharmacologic evidence that EHMT1 and EHMT2 are epigenetic regulators involved in γ-globin repression and represent a novel therapeutic target for SCD.
Introduction
Fetal hemoglobin (HbF, α2γ2) is the major hemoglobin during fetal life and is replaced by adult hemoglobin (HbA1, α2β2) during infancy.1 HbF induction is a well-validated strategy for sickle cell disease (SCD) treatment.2 HbF inhibits the polymerization of sickle hemoglobin (α2βS2) under deoxygenated conditions, and epidemiological studies indicate that HbF is a major modifier of the frequency of pain crises and mortality in SCD.2-4 Hydroxyurea, an HbF-inducing agent, is the only drug approved by the U.S. Food and Drug Administration for SCD treatment, but it has variable efficacy and dose-limiting myelosuppression.2,4 In addition to hydroxyurea, known activators of HbF include DNA hypomethylating agents, such as decitabine and 5-azacytidine,5,6 and histone deacetylase (HDAC) inhibitors,7,8 but achievement of therapeutic efficacy without significant cytotoxicity has limited their clinical applicability to date.2 Accordingly, more effective and safer HbF inducers are warranted.
The developmental transcriptional switch from fetal γ-globin to adult β-globin genes involves multiple cis-acting sequences and transcriptional factors, including BCL11A, GATA1, KLF1, and NFE2, acting as multiprotein complexes.9-11 The silencing of HbF in adults is also mediated by epigenetic changes, including DNA methylation and histone modifications, and several chromatin-modifying enzymes, including DNA methyltransferases,12 HDACs,8 PRMT5,13 and LSD1,12,14 have been shown to contribute to this process. However, it is likely that other epigenetic regulators remain to be identified.
Epigenetic regulators are emerging as attractive drug targets in various human diseases. The recent discovery of a large number of mutations in chromatin-modifying enzymes in cancer genomes15 has prompted the development of numerous small molecules targeting specific enzymes or proteins involved in the epigenetic regulation of gene expression, such as EZH2 inhibitors and bromodomain and extraterminal domain inhibitors.16 We used these recently developed molecules as tools to identify novel epigenetic regulators of γ-globin expression that are candidate therapeutic targets for the treatment of SCD.
Methods
Cell culture
Cryopreserved primary human CD34+ hematopoietic stem and progenitor cells (HSPCs) were obtained from Fred Hutchinson Cancer Research Center (Seattle, WA). The CD34+ cells were isolated from the peripheral blood of healthy donors after mobilization by administration of granulocyte colony-stimulating factor. Umbilical cord blood (CB) was harvested at Brigham and Women’s Hospital under an institutional review board–approved protocol, and CD34+ cells were purified using the MACS microbeads (Miltenyi Biotec). Cells were differentiated in vitro toward the erythroid lineage using a 3-phase culture method, as described previously.17 Murine erythroleukemia (MEL) cells (ATCC) were cultured in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum (FBS), 2% penicillin-streptomycin, and 1% l-glutamine (Life Technologies) at 37°C with 5% CO2.
Drug treatment of differentiating human cells and MEL cells
All compounds were purchased from Sigma Aldrich. UNC0638, entinostat (MS-275), decitabine dissolved in dimethylsulfoxide at 10 mM, or hydroxyurea dissolved in water at 10 mM were diluted in fresh culture media at the indicated final concentrations and durations. Culture medium containing 0.002% of dimethylsulfoxide served as a vehicle control. Every 1 to 2 days, culture media were completely replaced with new media containing freshly added drugs.
Morphologic analysis
Cell morphology was examined by May-Grünwald-Giemsa (Sigma-Aldrich) staining of cytospins. Cells were imaged using a Nikon Eclipse E400 microscope and SPOT imaging system (Diagnostic Instruments).
Cell proliferation and viability
Cell number and viability were determined with a hemocytometer by trypan blue staining. As an assessment of cytotoxicity, cells were plated in a 96-well culture plate and treated with a wide range of drug concentrations. On the respective days, total cellular adenosine triphosphate content was measured using CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the protocol. Luminescence was assessed by a multimode detector DTX880 (Beckman Coulter).
Generation of Ehmt2 knockout clones
Genomic deletions were created in MEL cells using pairs of chimeric single guide RNAs (sgRNAs) as previously described.18,19 Briefly, sgRNA-specifying oligos were chosen using publicly available online tools (supplemental Table 1 and supplemental Figure 1; see the Blood Web site).20 Oligos were phosphorylated, annealed, and cloned into pSpCas9(BB) (pX330; Addgene plasmid ID: 42230) using a Golden Gate Assembly strategy. MEL cells (2 × 106) were electroporated with 5 μg of each pX330-sgRNA plasmid and 0.5 μg of pmaxGFP plasmid (Lonza) at 250 V for 5 ms. The top 3% of GFP-expressing cells were sorted 48 hours postelectroporation to enrich for deletion. Sorted cells were plated clonally at limiting dilution. Clones were screened for deletion by conventional polymerase chain reaction (PCR; supplemental Table 2). Identified monoallelic and biallelic deletion clones were validated for knockout by real-time quantitative reverse-transcription PCR (qRT-PCR) (supplemental Tables 3 and 4) and by Sanger sequencing (supplemental Figure 2).
Expression of EHMT1 or EHMT2 shRNAs by lentiviral vector
Lentiviral vectors expressing short hairpin RNAs (shRNAs) were obtained from the RNAi Consortium of the Broad Institute (supplemental Table 5). Lentiviral vectors were transfected together with plasmid encoding packaging proteins and VSV-G using TRANS-LTI (Mirrus) into 293T cells. The media was changed after 24 hours, and the viral supernatant was collected 48 hours after transfection. Primary human progenitor cells were transduced on day 4 of culture by spin infection in the presence of 2 μg/mL polybrene (Sigma-Aldrich). Puromycin (Life Technologies) was added to the culture media at 2 μg/mL from day 5 to 14 of culture to select for infected cells.
RNA isolation and qRT-PCR assays
The MultiMACS Separator/Column system (Miltenyi Biotec) was used to isolate messenger RNA (mRNA) and synthesize complementary DNA, according to the manufacturer’s instructions. The qRT-PCR analysis of human genes was performed using the TaqMan Gene Expression Master Mix (Life Technologies) and the following primer-probe sets from Life Technologies: GAPDH (Hs99999905_m1), EHMT1 (Hs00964325_m1), EHMT2 (Hs00198710_m1), HBA1/2 (Hs00361191_g1), HBB (Hs00747223_g1), and HBG1/2 (Hs00361131_g1). The qRT-PCR analysis of murine genes was performed using iQ SYBR Green Supermix (Bio-Rad) and the primers indicated in supplemental Table 3. Each qRT-PCR was performed in duplicate in a 384-well plate on an ABI Prism 7900 HT PCR instrument (Life Technologies). The mean threshold cycle (Ct) for each assay was used for further calculations. The expression of all target genes was normalized to the GAPDH control gene (ΔCt).
Chromatin immunoprecipitation and sequencing (ChIP-seq)
Quantitative ChIP-seq was performed using the Mint-ChIP system (P.V.G., manuscript submitted July 2015). Briefly, a T7-adapter was ligated to micrococcal nuclease-digested native chromatin, with a different barcode for each sample (index #1). Samples were pooled and split for parallel immunoprecipitation of H3K9Ac (Active Motif, 39917), H3K9Me2 (Abcam, ab1220) and total H3 (Abcam, ab1791) for normalization. We used 2 micrococcal nuclease concentrations and 20 000 cells per ChIP assay; mouse carrier chromatin was added to have the chromatin equivalent of 400 000 cells per ChIP assay. Following ChIP, linear amplification of DNA was achieved by in vitro transcription. The resulting RNA was reverse transcribed to yield complementary DNA with Illumina priming sequences on both ends. PCR was used to generate libraries, adding a second barcode (index #2) to identify ChIP assays. Libraries were subjected to paired-end sequencing on the Illumina HiSeq2500 or Illumina NextSeq500 instrument. Reads were demultiplexed by ChIP assay (index #2) and sample (index #1) and aligned to hg19 using the Burrows-Wheeler Aligner. Reads that were mapped to >2 locations (X0 >2) were removed. To ensure that each nucleosomal fragment is represented by no more than 1 read, we removed PCR duplicates and duplicates resulting from linear amplification. Internal normalization to total H3 reads was used to quantify signal intensities. Tracks were visualized using integrative genome viewer. Reads that overlapped with the locus control region (LCR) (chr11:5312730-5297141), HBG2 (chr11:5276011-5274421), HBG1 (chr11:5271087-5269502), HBD (chr11:5255858-5254059), or HBB (chr11:5248301-5246696) were counted and normalized to total H3 reads to compare signal between samples. The data set is available in the Gene Expression Omnibus database (accession number GSE71422).
RNA sequencing (RNA-seq)
Details regarding RNA-seq analysis are provided in the supplementary Methods.
Flow cytometry
For cell surface marker analysis, 106 cells were resuspended in 100 µL phosphate-buffered saline with 2% FBS and stained with the phycoerythrin-conjugated CD235a (BD Biosciences, 555570) and allophycocyanin-conjugated CD71 (BD Biosciences, 341029) antibodies for 30 minutes on ice. For analysis of cytoplasmic HbF and H3K9Me2 chromatin mark, 106 cells were fixed and permeabilized using the Fix and Perm kit (Invitrogen) according to the manufacturer’s protocol. During the permeabilization step, cells were stained with fluorescein isothiocyanate–conjugated HbF-specific antibody from the Human Fetal Hemoglobin Test Kit (Invitrogen) or with fluorescein isothiocyanate–conjugated H3K9Me2-specific antibody (Abcam, ab64491), respectively. Stained cells were washed once with phosphate-buffered saline/2% FBS before analysis on the FACSCanto II flow cytometer (BS Biosciences). Data analysis was performed with FlowJo software.
HbF quantitation by HPLC
Cells were lysed and analyzed for hemoglobin composition using a Tosoh G7 glycohemoglobin analyzer (Tosoh Bioscience). The system uses nonporous ion exchange, high-performance liquid chromatography (HPLC) to measure HbF as a percentage of the total amount of hemoglobin present in the sample.
Western blot and antibodies
Nuclear extracts were prepared with the Nuclear Complex Co-IP kit (Active Motif), lysed in Laemmli sample buffer and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis. Protein lysates were run on tris(hydroxymethyl)aminomethane-HCl, 1 mm Criterion Precast gels (4% to 15%) (Bio-Rad) at a constant voltage. Proteins were transferred onto Immobilon-P transfer membranes (Millipore) at 0.60 A for 2 hours. Before staining with primary antibodies, blots were blocked in 5% nonfat dry milk (Santa Cruz Biotechnology) in tris(hydroxymethyl)aminomethane-buffered saline with Tween 20 (Santa Cruz Biotechnology) for 1 hour. For protein detection, primary antibodies detecting EHMT1 (mouse monoclonal, 1:1000, ab41969; Abcam), EHMT2 (rabbit polyclonal, 1:1000, 07-551; Millipore), and lamin B1 (rabbit monoclonal, 1:1000, 12586; Cell Signaling) were used.
Statistical analysis
GraphPad Prism version 5.0c was used to perform statistical analysis. Comparison of quantitative variables was performed using the 2-tailed Student t test or 1-way analysis of variance (ANOVA).
Results
Identification of EHMT1/2 inhibitors as inducers of γ-globin expression
We screened a set of small molecules targeting epigenetic modifiers, assessing the effects of each compound on γ-globin expression, relative to β-globin, in primary human erythroid cells. Among the novel compounds tested (supplemental Table 6), only 2 of them, BIX0129421 and UNC0638,22 showed a consistent, dose-dependent increase in the γ/β-globin mRNA ratio (supplemental Figure 3) at concentrations that did not substantially decrease cell viability (supplemental Figure 4). These 2 drugs selectively inhibit the catalytic activity of the histone methyltransferases EHMT1 (also known as GLP or KMT1D) and EHMT2 (also known as G9a or KMT1C) by blocking substrate access to their SET (Su(var)3-9, Enhancer-of-zeste, Trithorax) domains.22 Both proteins are key histone methyltransferases in euchromatin for mono- and dimethylation of lysine 9 on histone H3 (H3K9Me1/2). H3K9Me2 is a well-established repressive chromatin mark.23-25 EHMT1 and EHMT2 form a heterodimeric complex, which is thought to be the functional H3K9 methyltransferase in vivo.26-29
EHMT1/2 pharmacologic inhibition or knockdown increases γ-globin gene expression and HbF synthesis
To explore the role of EHMT1/2 in γ-globin repression in primary adult CD34+ HSPC-derived human erythroid precursors, we used pharmacologic inhibition with UNC0638 and genetic targeting using RNA interference. We first assessed the expression levels of α- and β-globin genes in primary adult human erythroid cells with or without UNC0638 treatment using qRT-PCR. Expression of HBG1/2 genes, but not that of HBB and HBA1/2, was significantly altered by UNC0638 (Figure 1A). Consistent with this finding, UNC0638 significantly increased the relative expression of γ-globin genes (Figure 1B), HbF level (Figure 1C-D), and the percentage of cells expressing HbF (Figure 1E), in a dose-dependent manner. These experiments were repeated on samples from 3 different donors and provided similar results (data not shown).
UNC0638 increases human γ-globin and HbF expression and mouse embryonic β-globin gene expression in a dose-dependent manner. (A) Fold change in HBA1/2, HBB, and HBG1/2 mRNA levels relative to GAPDH in primary adult human erythroid cells at day 14 of erythroid differentiation and after 10 days of UNC0638 treatment (mean ± standard deviation [SD], n = 5-6 biological replicates). (B) Relative expression of γ-globin genes in primary adult human erythroid cells at day 14 of erythroid differentiation and after 10 days of UNC0638 treatment (mean ± SD, n = 5-6 biological replicates). (C) HbF levels assessed by HPLC in primary adult human erythroid cells at day 14 of erythroid differentiation (mean ± SD, n = 4-8 biological replicates). (D) Representative HPLC chromatograms showing HbF abundance. (E) Flow cytometry analysis for F-cells: representative histograms showing the percentage of adult human erythroid cells expressing HbF and the fluorescence intensity relative to the cell number at day 14 of erythroid differentiation (mean ± SD, n = 3-4 biological replicates). ***P < .001. (F) Relative expression levels of the mouse embryonic globin genes Hbb-εy and Hbb-βh1 in MEL cells after 48 hours of UNC0638 treatment and in Ehmt2 knockout MEL cells (mean ± SD, n = 3 biological replicates). *P < .05. Cl., clone.
UNC0638 increases human γ-globin and HbF expression and mouse embryonic β-globin gene expression in a dose-dependent manner. (A) Fold change in HBA1/2, HBB, and HBG1/2 mRNA levels relative to GAPDH in primary adult human erythroid cells at day 14 of erythroid differentiation and after 10 days of UNC0638 treatment (mean ± standard deviation [SD], n = 5-6 biological replicates). (B) Relative expression of γ-globin genes in primary adult human erythroid cells at day 14 of erythroid differentiation and after 10 days of UNC0638 treatment (mean ± SD, n = 5-6 biological replicates). (C) HbF levels assessed by HPLC in primary adult human erythroid cells at day 14 of erythroid differentiation (mean ± SD, n = 4-8 biological replicates). (D) Representative HPLC chromatograms showing HbF abundance. (E) Flow cytometry analysis for F-cells: representative histograms showing the percentage of adult human erythroid cells expressing HbF and the fluorescence intensity relative to the cell number at day 14 of erythroid differentiation (mean ± SD, n = 3-4 biological replicates). ***P < .001. (F) Relative expression levels of the mouse embryonic globin genes Hbb-εy and Hbb-βh1 in MEL cells after 48 hours of UNC0638 treatment and in Ehmt2 knockout MEL cells (mean ± SD, n = 3 biological replicates). *P < .05. Cl., clone.
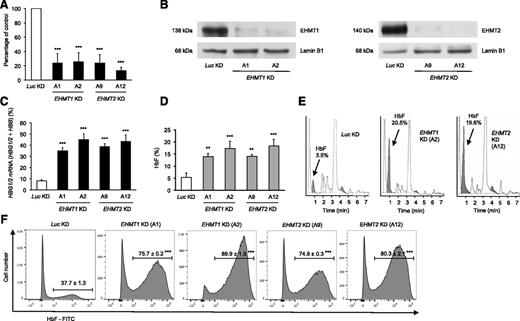
We validated the specificity of UNC0638 for EHMT1/2 by targeting EHMT1 and EHMT2 with lentiviral shRNAs in primary adult human erythroid cells. Knockdown of either EHMT1 or EHMT2 significantly increased the relative expression of γ-globin genes and the HbF level as compared with the control, up to 20% in some cases (Figure 2A-E). The percentage of cells expressing HbF was also significantly higher in cells with knockdown of EHMT1 or EHMT2 compared with control cells (Figure 2F). Knockdown of EHMT1 or EHMT2 did not significantly alter erythroid differentiation (supplemental Figure 5).
EHMT1 or EHMT2 knockdown increases γ-globin gene expression and HbF synthesis in primary adult human erythroid cells. (A) Validation of shRNA-mediated knockdown of EHMT1 or EHMT2 in primary adult human erythroid cells by qRT-PCR at day 14 of erythroid differentiation (mean ± SD, n = 5-6 biological replicates). (B) Validation of shRNA-mediated knockdown of EHMT1 or EHMT2 by western blot analysis in primary adult human erythroid cells at day 9 of erythroid differentiation and after 4 days of puromycin selection. shRNA-mediated knockdown of EHMT1 or EHMT2 increases γ-globin mRNA levels (mean ± SD, n = 5-6 biological replicates) (C) and HbF levels assessed by HPLC in primary adult human erythroid cells at day 14 of erythroid differentiation (mean ± SD, n = 4-6 biological replicates) (D). (E) Representative HPLC chromatograms showing HbF abundance. (F) Flow cytometry analysis for F cells: representative histograms showing the percentage of adult human erythroid cells expressing HbF and the fluorescence intensity relative to the cell number at day 14 of erythroid differentiation (mean ± SD, n = 3-5 biological replicates). **P < .01; ***P < .001.
EHMT1 or EHMT2 knockdown increases γ-globin gene expression and HbF synthesis in primary adult human erythroid cells. (A) Validation of shRNA-mediated knockdown of EHMT1 or EHMT2 in primary adult human erythroid cells by qRT-PCR at day 14 of erythroid differentiation (mean ± SD, n = 5-6 biological replicates). (B) Validation of shRNA-mediated knockdown of EHMT1 or EHMT2 by western blot analysis in primary adult human erythroid cells at day 9 of erythroid differentiation and after 4 days of puromycin selection. shRNA-mediated knockdown of EHMT1 or EHMT2 increases γ-globin mRNA levels (mean ± SD, n = 5-6 biological replicates) (C) and HbF levels assessed by HPLC in primary adult human erythroid cells at day 14 of erythroid differentiation (mean ± SD, n = 4-6 biological replicates) (D). (E) Representative HPLC chromatograms showing HbF abundance. (F) Flow cytometry analysis for F cells: representative histograms showing the percentage of adult human erythroid cells expressing HbF and the fluorescence intensity relative to the cell number at day 14 of erythroid differentiation (mean ± SD, n = 3-5 biological replicates). **P < .01; ***P < .001.
UNC0638 does not alter cellular morphology, proliferation, or erythroid differentiation
UNC0638 did not alter cellular morphology at a concentration of 0.25 μM and lower (Figure 3A). Cell proliferation and viability were not significantly affected up to 0.1 μM, regardless of the treatment duration (Figure 3B-C). After a 7-day treatment, UNC0638 50% inhibitory concentration was 1 μM, compared with 0.15 μM for entinostat, an inhibitor of HDAC1/2/3, and 0.08 μM for decitabine (supplemental Figure 6). As observed with EHMT1 or EHMT2 shRNA-mediated knockdown, EHMT1/2 inhibition by UNC0638 was not associated with a block in erythroid differentiation (Figure 3D), in contrast with entinostat and, to a lesser extent, decitabine treatment (supplemental Figure 7).
Effect of UNC0638 treatment on cell morphology, proliferation, and differentiation of primary adult human erythroid cells in culture. (A) Representative images of the morphology of primary adult human erythroid cells differentiated ex vivo in the presence of 0.25 μM UNC0638 or the vehicle control. Bar represents 10 μm. (B) Cell proliferation assessed by trypan blue staining in the presence of 3 doses of UNC0638 or the vehicle control (mean ± SD, n = 3 biological replicates). (C) Cell Titer Glo curves for a wide range of UNC0638 concentrations and for 3 different treatment duration (mean ± SD, n = 5-6 biological replicates). (D) Flow cytometry analysis of cell-surface differentiation marker expression at day 15 of erythroid differentiation (mean ± SD, n = 3-4 biological replicates). *P < .05; **P < .01.
Effect of UNC0638 treatment on cell morphology, proliferation, and differentiation of primary adult human erythroid cells in culture. (A) Representative images of the morphology of primary adult human erythroid cells differentiated ex vivo in the presence of 0.25 μM UNC0638 or the vehicle control. Bar represents 10 μm. (B) Cell proliferation assessed by trypan blue staining in the presence of 3 doses of UNC0638 or the vehicle control (mean ± SD, n = 3 biological replicates). (C) Cell Titer Glo curves for a wide range of UNC0638 concentrations and for 3 different treatment duration (mean ± SD, n = 5-6 biological replicates). (D) Flow cytometry analysis of cell-surface differentiation marker expression at day 15 of erythroid differentiation (mean ± SD, n = 3-4 biological replicates). *P < .05; **P < .01.
UNC0638 or Ehmt2 knockout increases mouse embryonic β-globin gene expression
To validate the capacity of EHMT1/2 inhibitors to modulate the transcription of β-globin genes in an independent system, we tested UNC0638 in MEL cells. These cells are transformed erythroid precursors blocked at about the proerythroblast stage of differentiation and serve as a cellular model to study mouse erythropoiesis and globin switching.30,31 In these cells, UNC0638 treatment led to a relative increase in Hbb-εy and, to a lesser extent, in Hbb-βh1 transcript level, in a dose-dependent manner (Figure 1F). In parallel with pharmacologic inhibition, we used CRISPR/Cas9 gene editing in MEL cells to generate clones with biallelic inactivation of Ehmt2. Ehmt2 knockout clones harbored a marked increase in Hbb-εy and Hbb-βh1 expression compared with parental cells, at levels comparable to those achieved with 1 μM UNC0638 (Figure 1F). We were unable to generate clones with homozygous inactivation of Ehmt1, despite extensive efforts, suggesting that this enzyme may be essential in MEL cells.
H3K9Me2 levels decrease during erythroid differentiation and are further decreased by EHMT1/2 pharmacologic inhibition or knockdown
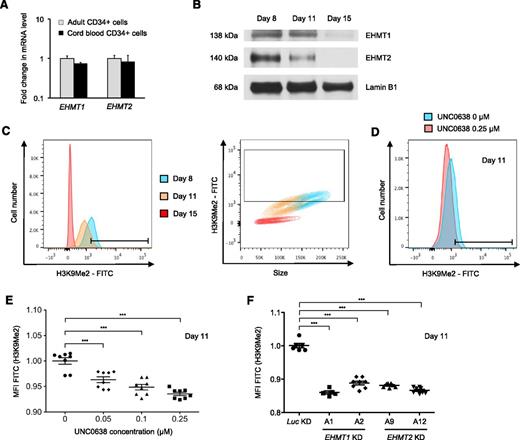
We next examined the level of EHMT1 and EHMT2 expression during the course of erythroid differentiation. Adult or CB CD34+ cells had similar EHMT1 and EHMT2 transcript levels (Figure 4A), indicating that EHMT1/2 expression itself is unlikely to be the primary mediator of developmental silencing. Western blot analysis showed that the protein levels of both EHMT1 and EHMT2 decreased between proerythroblasts (day 8 of erythroid differentiation) and more mature erythroblasts (days 11 and 15 of erythroid differentiation) (Figure 4B). Because the EHMT1/2 complex catalyzes mono- and dimethylation of H3K9, we investigated whether EHMT1/2 inhibition or knockdown decreases the global level of H3K9Me2 chromatin mark in primary adult human erythroid cells using flow cytometry. Consistent with EHMT1 and EHMT2 protein levels, global H3K9Me2 levels progressively decreased during erythroid differentiation, in parallel with the reduction in cell size, and became undetectable at day 15 of erythroid differentiation (Figure 4C). As expected, UNC0638 treatment caused a dose-dependent reduction in H3K9Me2 (Figure 4D-E). Similarly, shRNA-mediated knockdown of EHMT1 or EHMT2 decreased global H3K9Me2 levels in erythroid precursors (Figure 4F).
H3K9Me2 levels decrease during erythroid differentiation and are further decreased by EHMT1/2 pharmacologic inhibition or knockdown in primary adult human erythroid cells. (A) EHMT1 and EHMT2 mRNA expression in human CD34+ cells from adult or umbilical CB. Results show the mean value ± SD of 2 different donors for adult CD34+ and 2 pools of 10 cords for CB CD34+. (B) Western blot analysis showing EHMT1, EHMT2, and lamin B1 (loading control) in primary adult human cells during erythroid differentiation ex vivo. (C) Representative flow plots of H3K9Me2 level according to the cell number and the size during erythroid differentiation ex vivo. (D) Representative histogram showing the fluorescence intensity for H3K9Me2 relative to the cell number in erythroid cells differentiated in the presence of 0.25 μM UNC0638 or the vehicle control for 7 days. (E) Dot plots showing the median fluorescence intensity (MFI) for H3K9Me2 after 7 days of treatment according to the UNC0638 concentration (mean ± SD, n = 8 biological replicates). (F) Dot plots showing the MFI for H3K9Me2 in EHMT1 or EHMT2 knockdown cells (mean ± SD, n = 5-8 biological replicates). ***P < .001.
H3K9Me2 levels decrease during erythroid differentiation and are further decreased by EHMT1/2 pharmacologic inhibition or knockdown in primary adult human erythroid cells. (A) EHMT1 and EHMT2 mRNA expression in human CD34+ cells from adult or umbilical CB. Results show the mean value ± SD of 2 different donors for adult CD34+ and 2 pools of 10 cords for CB CD34+. (B) Western blot analysis showing EHMT1, EHMT2, and lamin B1 (loading control) in primary adult human cells during erythroid differentiation ex vivo. (C) Representative flow plots of H3K9Me2 level according to the cell number and the size during erythroid differentiation ex vivo. (D) Representative histogram showing the fluorescence intensity for H3K9Me2 relative to the cell number in erythroid cells differentiated in the presence of 0.25 μM UNC0638 or the vehicle control for 7 days. (E) Dot plots showing the median fluorescence intensity (MFI) for H3K9Me2 after 7 days of treatment according to the UNC0638 concentration (mean ± SD, n = 8 biological replicates). (F) Dot plots showing the MFI for H3K9Me2 in EHMT1 or EHMT2 knockdown cells (mean ± SD, n = 5-8 biological replicates). ***P < .001.
UNC0638 decreases H3K9Me2 and increases H3K9Ac at the β-globin locus
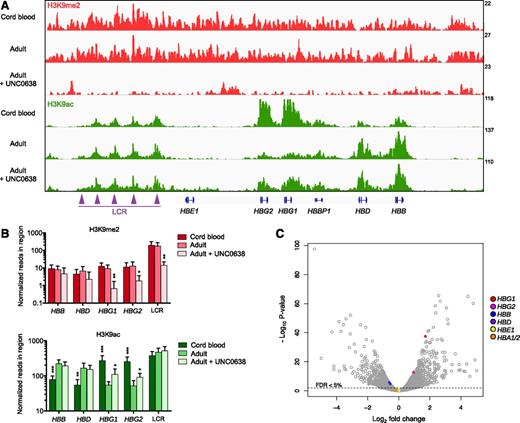
To examine the impact of UNC0638 on chromatin state, we performed ChIP-seq analysis in primary human adult erythroblasts after 11 days of erythroid differentiation in the presence or absence of the compound. We focused on 2 chromatin marks, the repressive H3K9Me2 mark and the antagonist activating H3K9Ac mark. UNC0638 treatment significantly reduced H3K9Me2 levels, genome wide, at the 2 γ-globin genes, and at the LCR (supplemental Figure 8 and Figure 5A-B). The depletion in H3K9Me2 induced by UNC0638 was associated with a concomitant increase in the antagonist chromatin mark H3K9Ac, which was especially pronounced at the γ-globin gene region (Figure 5A-B). In addition, we examined the chromatin state of primary human erythroblasts derived from CB CD34+. The abundance of H3K9Me2 did not differ between CB and adult erythroblasts, genome wide (supplemental Figure 8) and at the β-globin locus (Figure 5A-B).
Effects of UNC0638 on H3K9Me2 and H3K9Ac chromatin occupancy at the β-globin locus and gene expression in primary human erythroblasts. (A) Integrative genome viewer screenshot at the β-globin locus in erythroblasts derived from umbilical CB or adult CD34+ cells that were differentiated in the presence of 0.25 μM UNC0638 or the vehicle control. ChIP-seq analysis was performed at day 11 of erythroid differentiation and after 7 days of treatment. H3K9Me2 or H3K9Ac reads are normalized to H3 reads for Mint-ChIP internal normalization, as indicated by the values on the y-axis. The tracks represent the pool of 3 biological replicates. (B) Corresponding histograms representing the normalized number of reads in the indicated region of the β-globin locus for both histone marks. H3K9Me2 or H3K9Ac reads are normalized to H3 reads for Mint-ChIP internal normalization (mean ± SD, n = 3 biological replicates). *P < .05; **P < .01; ***P < .001, relative to untreated adult erythroblasts. (C) Volcano plot illustrating changes in gene expression induced by UNC0638. RNA-seq analysis was performed in primary adult human erythroid cells at day 11 of erythroid differentiation and after 7 days of treatment with 0.25 μM UNC0638 or the vehicle control. The plot represents statistical significance vs the fold change in gene expression between the 2 conditions. Results from 3 biological replicates are shown. FDR, false discovery rate.
Effects of UNC0638 on H3K9Me2 and H3K9Ac chromatin occupancy at the β-globin locus and gene expression in primary human erythroblasts. (A) Integrative genome viewer screenshot at the β-globin locus in erythroblasts derived from umbilical CB or adult CD34+ cells that were differentiated in the presence of 0.25 μM UNC0638 or the vehicle control. ChIP-seq analysis was performed at day 11 of erythroid differentiation and after 7 days of treatment. H3K9Me2 or H3K9Ac reads are normalized to H3 reads for Mint-ChIP internal normalization, as indicated by the values on the y-axis. The tracks represent the pool of 3 biological replicates. (B) Corresponding histograms representing the normalized number of reads in the indicated region of the β-globin locus for both histone marks. H3K9Me2 or H3K9Ac reads are normalized to H3 reads for Mint-ChIP internal normalization (mean ± SD, n = 3 biological replicates). *P < .05; **P < .01; ***P < .001, relative to untreated adult erythroblasts. (C) Volcano plot illustrating changes in gene expression induced by UNC0638. RNA-seq analysis was performed in primary adult human erythroid cells at day 11 of erythroid differentiation and after 7 days of treatment with 0.25 μM UNC0638 or the vehicle control. The plot represents statistical significance vs the fold change in gene expression between the 2 conditions. Results from 3 biological replicates are shown. FDR, false discovery rate.
In contrast with the β-globin locus, the α-globin locus had low basal levels of H3K9Me2 that were not significantly altered by UNC0638 treatment (supplemental Figure 9), and no change in H3K9Ac levels at the HBA1 and HBA2 loci was observed following UNC0638 treatment. H3K9Me2 and H3K9Ac abundance at other loci, including the control gene GAPDH and the main erythroid transcription factors, such as GATA1, KLF1, and NFE2, were not significantly affected by UNC0638 treatment (supplemental Figures 10-12), consistent with our data showing that EHMT1/2 inhibition does not affect erythroid differentiation. H3K9Me2 levels at the γ-globin repressors BCL11A and MYB loci were significantly reduced after UNC0638 treatment, with a slight increase in H3K9Ac levels (supplemental Figure 12). Overall, these data demonstrate that UNC0638 decreases H3K9Me2 and increases H3K9Ac at the β-globin gene cluster but does not significantly alter these histone marks at the α-globin locus, consistent with the specific derepression of γ-globin gene expression.
Effects of UNC0638 on global gene expression
To evaluate the effects of UNC0638 treatment on global gene expression, we performed RNA-seq analysis using CD34+ HSPCs from the same donor and culture conditions as for ChIP-seq analysis. Overall, 2914 genes had significantly altered expression (FDR <0.05) following treatment with UNC0638 (Figure 5C). Among genes with at least a twofold change, 387 genes were significantly upregulated and 252 downregulated. HBG1 and, to a lesser extent, HBG2 were among the most significantly upregulated genes by UNC0638. Expression of HBB and HBD were modestly but significantly decreased, whereas HBE1 expression remained stable. HBA1 and HBA2 expression were not significantly altered (Figure 5C; Table 1), consistent with our qRT-PCR data (Figure 1A). Altogether, the changes in α- and β-globin gene expression induced by UNC0638 reflected the changes observed by ChIP-seq (supplemental Figure 13). The expression of the major erythroid transcription factors, GATA1, KLF1, and NFE2, was unaffected by UNC0638, consistent with our finding that UNC0638 does not significantly alter erythroid differentiation. The γ-globin repressors BCL11A and MYB were not significantly deregulated by UNC0638 (Table 1), indicating that EHMT1/2 inhibition does not upregulate γ-globin genes through downregulation of BCL11A or MYB.
We used gene set enrichment analysis32 to identify gene expression patterns associated with EHMT1/2 inhibition in gene sets of interest. The highest enrichment score associated with UNC0638 treatment was observed for the heme metabolism signature (supplemental Figure 14), which supports the fact that EHMT1/2 inhibition promotes HbF synthesis. Similarly, a GATA1 signature was significantly enriched (supplemental Figure 15).
EHMT1/2 inhibition or knockdown shows additive effects with entinostat or decitabine
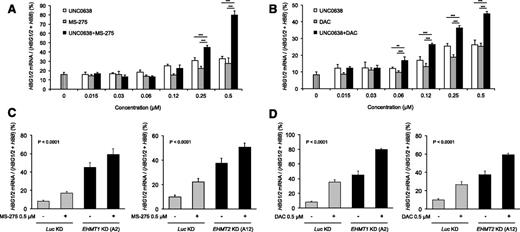
In order to achieve even higher induction of HbF, we investigated the effects of combinatorial EHMT1/2 inhibition with HDAC inhibitors and DNA hypomethylating agents, other epigenetic drugs that induce γ-globin gene expression. The combination of UNC0638 and entinostat, an HDAC inhibitor, had additive effects on the induction of γ-globin gene expression (Figure 6A). The combination of UNC0638 with decitabine, a DNA hypomethylating agent, also had additive effects (Figure 6B). Similarly, additive effects were observed in EHMT1 or EHMT2 knockdown cells treated with 0.5 μM entinostat (Figure 6C) or decitabine (Figure 6D). These data suggest that EHMT1/2 inhibition induces γ-globin expression through a mechanism of action distinct from entinostat or decitabine, and that combination therapy has the potential for high-level HbF induction.
Combination of EHMT1/2 pharmacologic inhibition or knockdown with entinostat or decitabine shows additive effects on γ-globin gene induction in primary adult human erythroid cells. (A) Histograms showing the relative percentage of γ-globin genes expression according to the concentration of drug(s). qRT-PCR analysis was performed at day 14 of erythroid differentiation, after treatment with UNC0638 only, entinostat (MS-275) only, or UNC0638 + MS-275 for 7 days (from day 7 to day 14) (mean ± SD, n = 5 biological replicates). ***P < .001. (B) Histograms showing the relative percentage of γ-globin genes expression according to the concentration of drug(s). qRT-PCR analysis was performed at day 14 of erythroid differentiation, after treatment with UNC0638 only (from day 7 to day 14), decitabine (DAC) only (from day 11 to day 14), or UNC0638 + DAC (from day 7 to day 14 for UNC0638 and from day 11 to day 14 for DAC) (mean ± SD, n = 5 biological replicates). **P < .01; ***P < .001. (C) Histograms showing the relative percentage of γ-globin genes expression in knockdown cells, in the presence of 0.5 μM MS-275 or the vehicle control for 3 days (from day 11 to day 14). qRT-PCR analysis was performed at day 14 of erythroid differentiation (mean ± SD, n = 4-6 biological replicates). One-way ANOVA, P < .001. (D) Histograms showing the relative percentage of γ-globin genes expression in knockdown cells, in the presence of 0.5 μM DAC or the vehicle control for 3 days (from day 11 to day 14). qRT-PCR analysis was performed at day 14 of erythroid differentiation (mean ± SD, n = 4-6 biological replicates). One-way ANOVA, P < .001.
Combination of EHMT1/2 pharmacologic inhibition or knockdown with entinostat or decitabine shows additive effects on γ-globin gene induction in primary adult human erythroid cells. (A) Histograms showing the relative percentage of γ-globin genes expression according to the concentration of drug(s). qRT-PCR analysis was performed at day 14 of erythroid differentiation, after treatment with UNC0638 only, entinostat (MS-275) only, or UNC0638 + MS-275 for 7 days (from day 7 to day 14) (mean ± SD, n = 5 biological replicates). ***P < .001. (B) Histograms showing the relative percentage of γ-globin genes expression according to the concentration of drug(s). qRT-PCR analysis was performed at day 14 of erythroid differentiation, after treatment with UNC0638 only (from day 7 to day 14), decitabine (DAC) only (from day 11 to day 14), or UNC0638 + DAC (from day 7 to day 14 for UNC0638 and from day 11 to day 14 for DAC) (mean ± SD, n = 5 biological replicates). **P < .01; ***P < .001. (C) Histograms showing the relative percentage of γ-globin genes expression in knockdown cells, in the presence of 0.5 μM MS-275 or the vehicle control for 3 days (from day 11 to day 14). qRT-PCR analysis was performed at day 14 of erythroid differentiation (mean ± SD, n = 4-6 biological replicates). One-way ANOVA, P < .001. (D) Histograms showing the relative percentage of γ-globin genes expression in knockdown cells, in the presence of 0.5 μM DAC or the vehicle control for 3 days (from day 11 to day 14). qRT-PCR analysis was performed at day 14 of erythroid differentiation (mean ± SD, n = 4-6 biological replicates). One-way ANOVA, P < .001.
Discussion
Our study demonstrates that EHMT1 and EHMT2 repress γ-globin gene expression and are potential therapeutic targets for SCD. We found that UNC0638, a compound that selectively inhibits the EHMT1/2 complex, induced γ-globin gene expression and HbF synthesis in primary human adult erythroid cells with low cytotoxicity and without inhibiting erythroid differentiation. Our data also suggest that EHMT1 and EHMT2 have distinct patterns of activity compared with known regulators of the β-globin switch, such as BCL11A and MYB. We demonstrated that genetic targeting of EHMT1 or EHMT2 using lentiviral shRNAs also induced γ-globin gene expression and HbF synthesis in primary human erythroid cells. Similarly, we showed that UNC0638 treatment and Ehmt2 CRISPR/Cas9-mediated knockout in the murine cell line MEL increased the expression of both Hbb-εy and Hbb-βh1 embryonic globin genes.
Recent studies have also indicated that EHMT1 and EHMT2 are involved in the regulation of γ-globin gene expression. In a gene expression profiling study in CD34+ cells following treatment with UNC0638, HBG1 and HBG2 were among the most significantly upregulated genes,33 in accordance with our RNA-seq data. An independent study performed contemporaneously to ours showed that EHMT2 inhibition by UNC0638 stimulated HbF production by facilitating LDB1 complex occupancy at the γ-globin promoters and LCR/γ-globin looping.34 Chaturvedi et al previously reported a role for Ehmt2 in the maintenance of β-globin gene transcription in MEL cells.30,35 In aggregate, our own and previous data converged on a critical role for EHMT1/2 in γ-globin repression in adult erythroid cells.
Treatment with UNC0638 caused genome-wide depletion of H3K9Me2 upon EHMT1/2 inhibition in primary human adult erythroid cells, consistent with previous studies,21,22,33 and revealed that this depletion was particularly significant at the γ-globin genes and the LCR of the β-globin locus, thereby facilitating γ-globin gene transcription through increasing chromatin accessibility.36 How H3K9Me2 marks are established on the β-globin locus in human hematopoietic cells remains to be clarified. In MEL cells, Ehmt2 is recruited to the β-globin locus in an Nf-E2-dependent manner.30 EHMT2 has also been shown to be recruited by UHRF1, which binds methylated cytosine guanine dinucleotides and recruits the transcriptional repressors DNMT1 and HDAC1.37
Previous studies showed that Ehmt2 exhibits a complex role in the regulation of mouse β-globin gene transcription, repressing Hbb-εy while activating β-Major, independently from its methyltransferase activity.30,35,38 In primary human adult erythroid cells, increased EHMT2 occupancy at the γ-globin gene promoters has been observed following UNC0638 treatment, suggesting that EHMT2 may have a positive role in γ-globin expression.34 However, the functional effects of EHMT2 binding to the γ-globin locus are unclear.
H3K9 acetylation and methylation are mutually exclusive.39 Our ChIP-seq analysis demonstrated that depletion in H3K9Me2 following treatment with UNC0638 was associated with a concomitant increase in H3K9Ac at the γ-globin gene region. Consistent with these epigenetic changes, we identified that the combination of UNC0638 with an HDAC inhibitor, entinostat, increased γ-globin expression more than either compound alone. UNC0638 and decitabine also had additive effects on γ-globin gene expression. Importantly, these findings suggest that combination therapy targeting multiple epigenetic marks has the potential to induce dramatic and clinically significant induction of HbF production. Given the genome-wide effects of UNC0638, it is likely that EHMT1/2 inhibition also enhances the effect of both HDAC inhibitors and DNA hypomethylating agents independently at other genomic regions, including tumor suppressor genes loci. Therefore, these combined therapies might also provide clinical benefit in hematologic malignancies, such as myelodysplastic syndromes, in which hypomethylating agents have been shown to be beneficial.40
Although UNC0638 is a potent chemical tool for in vitro studies, this compound has poor pharmacokinetic properties.22 Development of EHMT1/2 inhibitors with improved pharmacokinetic properties will enable in vivo validation studies and possible clinical application. Clinical application of EHMT2 inhibition has also been suggested for acute myeloid leukemia, as a means to counteract the proliferation and self-renewal of acute myeloid leukemia cells by attenuating HOXA9-dependent transcription.41 Furthermore, a recent study using CRISPR/Cas9 screening of chromatin regulatory protein domains identified Ehmt1 and Ehmt2 among the top genes associated with leukemia dependencies.42
Our data provide genetic and pharmacologic evidence that EHMT1/2 inhibition has the ability to induce γ-globin expression and HbF synthesis in primary adult human erythroid cells and deserves further investigation as a novel therapeutic approach for the treatment of SCD and β-hemoglobinopathies.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE71422).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a Clinical Research Award from the Doris Duke Charitable Foundation; the National Institutes of Health National Heart, Lung, and Blood Institute (1U01HL117720); and a Leukemia and Lymphoma Society Scholar Award (B.L.E.). Enriched CD34+ cells were obtained through the Core Center of Excellence program with partial funding support by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (grant DK56465). A.R. received partial financial support from the French Society of Hematology (Société Française d’Hématologie). M.C.C. was supported by a NIDDK Award (F30DK103359-01A1).
Authorship
Contribution: A.R., P.V.G., M.C.C., M.M., and D.M.D. performed experiments; A.R., P.V.G., M.C.C., and J.M.K.-B. analyzed data; E.B.H. provided samples; B.E.B., S.H.O., and D.E.B. contributed to the design of the project; P.V.G. and M.C.C. contributed to writing the manuscript; A.R. and B.L.E. designed research and wrote the manuscript; B.L.E. conceptualized the idea and supervised the project; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Benjamin L. Ebert, 1 Blackfan Circle, Karp 5.210, Boston, MA 02115; e-mail: Benjamin_Ebert@dfci.harvard.edu.

![Figure 1. UNC0638 increases human γ-globin and HbF expression and mouse embryonic β-globin gene expression in a dose-dependent manner. (A) Fold change in HBA1/2, HBB, and HBG1/2 mRNA levels relative to GAPDH in primary adult human erythroid cells at day 14 of erythroid differentiation and after 10 days of UNC0638 treatment (mean ± standard deviation [SD], n = 5-6 biological replicates). (B) Relative expression of γ-globin genes in primary adult human erythroid cells at day 14 of erythroid differentiation and after 10 days of UNC0638 treatment (mean ± SD, n = 5-6 biological replicates). (C) HbF levels assessed by HPLC in primary adult human erythroid cells at day 14 of erythroid differentiation (mean ± SD, n = 4-8 biological replicates). (D) Representative HPLC chromatograms showing HbF abundance. (E) Flow cytometry analysis for F-cells: representative histograms showing the percentage of adult human erythroid cells expressing HbF and the fluorescence intensity relative to the cell number at day 14 of erythroid differentiation (mean ± SD, n = 3-4 biological replicates). ***P < .001. (F) Relative expression levels of the mouse embryonic globin genes Hbb-εy and Hbb-βh1 in MEL cells after 48 hours of UNC0638 treatment and in Ehmt2 knockout MEL cells (mean ± SD, n = 3 biological replicates). *P < .05. Cl., clone.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/16/10.1182_blood-2015-06-649087/4/m_1930f1.jpeg?Expires=1769388889&Signature=YP55IFRF08NpphebxwKyQG2DMZF2kvgaAXOMk03eZBD4WcPbwRdtfat~8qWdCHQYir56dapUhRLwi4NdbsxV3U6XAIa1spuqO4gkJgkLjeIYnyMvtdxnfW-HJCBeJQPQabmQvSOVdfY9HkdaNKJib0QCLGMfCu8tpRxGXECfvsYF4luq5zKUPA~mi4n2snAwPB5fih9YQDOJ3SrZZF8rVeoZs9CzvYBHvnahqxLYMlA9dyGUUDHhJoqgKmozHDDo~cnA8KatOeSKDo9tnr4QUwF4oM0zW9EV-iiMalCvcLq~wiDYQXYR8eIPpQVjgKV7RP5oh8LRpJEAdDdzz61TQw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)