Abstract
Introduction - With rapidly advancing sequencing technology, the extent of genetic diversity in AML has never been more apparent. A Òone size fits allÓ approach can no longer be justified. Sequencing studies have also uncovered several actionable targets, yet no targeted therapies are FDA approved for use in the US. AML therefore, has significant potential for personalized therapeutics. Several challenges exist, however. The masses of data generated by high-throughput technologies are challenging to manage, visualize, and convert to knowledge required to improve outcomes. A cross-disciplinary systems biology effort is required, to visualize inter-connected events within leukemic blasts that ultimately contribute to the disease phenotype and inform on rational selection of therapeutic approaches. In the current study, we outline a complimentary functional and genomic screening approach to identify clinical drug candidates for re-purposing in patients with relapsed refractory AML.
Methods - In this proof-of principle study, we optimized an ex vivo high-throughput drug screening platform measuring AML cell survival after exposure to over 200 U.S. FDA approved oncology drugs (including conventional chemotherapeutics, proteasome inhibitors, anti-metabolites, transcriptional inhibitors and targeted kinase inhibitors). This multiplex assay is designed for individual AML patients and tests agents over a 10,000-fold concentration range. We screened patient derived blasts against normal bone marrow mononuclear cells to identify the most effective leukemia selective agents. To compliment this functional ex-vivo screen, we employed a genomics approach using predictive simulation software to generate patient specific avatars which map individual dysregulated and interconnecting signaling pathways. The avatar technology will then identify candidate agents at critical impact points within these pathways.
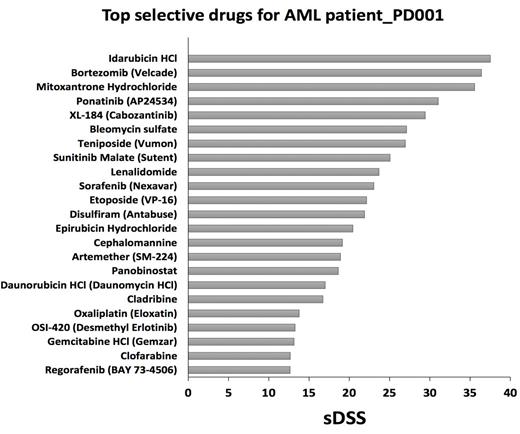
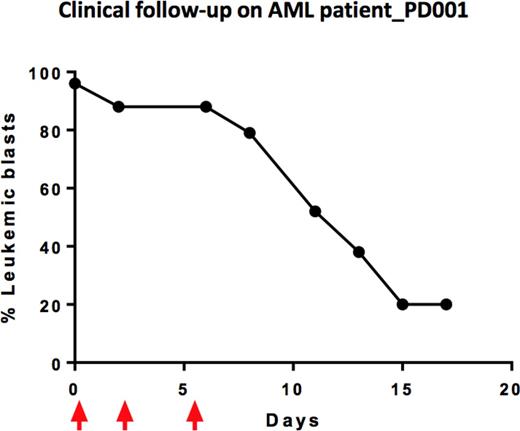
Results - Through phenotype screening of primary cells collected from a highly refractory AML patient (patient # PD001), our ex vivo assay identified a list of drugs based on their ability to effectively and selectively reduce the viability of the patients leukemic blasts in culture. Candidate agents are listed according to selective drug sensitivity scores (sDSS), calculated based on the comparative ability of each drug to reduce viability of primary cells vs normal bone marrow mononuclear cells. The highest sDSS indicates the most selective and effective drugs for each individual patient (Figure 1, patient sample PD001). The patient specific avatar generated for patient PD001 identified a series of dysregulated pathways converging on cell proliferation and viability. Both functional and genomic approaches identified the tyrosine kinase inhibitor ponatinib, as a potentially relevant clinical candidate. Potentially effective combination approaches were also predicted (e.g. ponatinib with rosuvastatin, ponatinib with decitabine). Since our ex vivo assay identified bortezomib (Velcade) as a clinical candidate and since we successfully negotiated off-label use of this agent, we selected Velcade for a therapeutic trial in patient PD001. After three doses of Velcade at 1.5 mg/m2 (Days 1,4 and 8), serial blood counts revealed a dramatic fall in total white count and circulating blasts (96% to 20%) (Figure 2). This is noteworthy since single agent Velcade is generally not capable of producing clinically meaningful responses for patients with refractory AML.
Conclusions - Our study justifies continued development of this novel, iterative functional/genomics approach to personalized therapeutics in AML. Our model identifies candidate drugs that can be readily re-purposed for immediate clinical use, whilst at the same time providing insights into underlying mechanism of action, informing on rationally designed combination strategies and biomarker candidates.
Ex vivo drug screening results from refractory patient_PD001 identifies the proteasome inhibitor bortezomib (Velcade) as one of the top selective and effective drugs. The higher the sDSS, the more effective and selective the drug is.
Ex vivo drug screening results from refractory patient_PD001 identifies the proteasome inhibitor bortezomib (Velcade) as one of the top selective and effective drugs. The higher the sDSS, the more effective and selective the drug is.
Clinical follow-up of AML patient-PD001 shows chemosensitivity to bortezomib (Velcade). Arrows arrows indicate dosing. Velcade decreased the percentage of leukemic blasts from 96% to 20%.
Clinical follow-up of AML patient-PD001 shows chemosensitivity to bortezomib (Velcade). Arrows arrows indicate dosing. Velcade decreased the percentage of leukemic blasts from 96% to 20%.
Vega:Seatle Genetics: Honoraria; NIH: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.