Abstract
Donor telomere length was recently shown to be associated with survival in severe aplastic anemia patients [1] who undergo unrelated donor HCT. We hypothesize that telomere length may also be clinically useful in selecting donors for acute leukemia patients who undergo HCT. To that end, we are developing predictive models, based on leukocyte telomere length together with clinical variables as inputs, and overall survival as the clinical outcome. The models classify donors as either "preferred" or "not preferred," to provide a decision making tool that can be used to select donors when multiple donors are available. We describe development of the classifier models using survival as the primary outcome and explore the predictive ability of the models for relapse and treatment-related mortality.
Whole blood samples and clinical information were obtained from the Center for International Blood and Marrow Transplant Research (CIBMTR) for 300 acute leukemia patients who underwent unrelated donor bone marrow or peripheral blood stem cell transplantation, 2004-2008. Inclusion criteria for adult and pediatric patients included: acute myeloid leukemia (AML) and acute lymphocytic leukemia (ALL) of any stage; 8/8 HLA-matched; and myeloablative conditioning. The samples were processed in Telomere Diagnostic's Clinical Laboratory Improvement Amendments (CLIA) certified laboratory, using a qPCR method to obtain relative leukocyte telomere length from the donor blood samples.
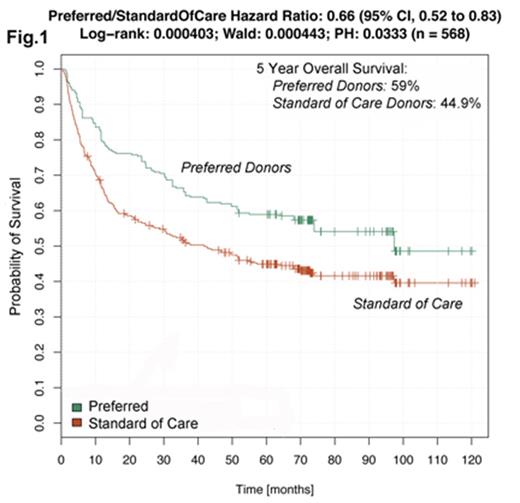
We used a multivariate analysis looking at donor telomere length and 30 other variables to identify a subset of donors that resulted in improved outcomes in the respective patients. With this approach, we derived an algorithm that identified 268 patients with substantially improved overall survival ("preferred group") compared to the entire study group ("standard of care group"). Preferred cases identified by the algorithm exhibited an improvement in median survival of 55.7 months vs standard of care (97 mos. median survival for preferred, vs. 41.3 mos. for standard of care, as shown in Fig. 1. Five-year overall survival was 59% for the preferred group, and 44.9% for the standard of care group.
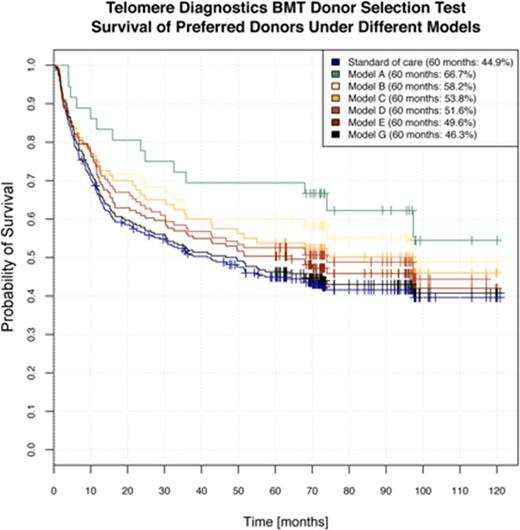
We performed additional analysis to create multivariate predictive models, which we call "classifiers," for donor selection. Rather than creating just a single classifier, we created six classifiers that provide a range of stringency levels (13%-80%). Classifiers with high stringency yield a smaller fraction of preferred donors to non-preferred donors; classifiers with low stringency yield a higher fraction of preferred donors to non-preferred donors. Using this graded approach makes differences in donor quality apparent, even where the recipient has few potential donors from which to choose. However, this type of analysis does not yield a continuous donor score or allow direct quantification of the significance of a variable in the model.
To quantify the utility of the six classifiers, we estimated performance using repeated 10-fold cross-validation [2]. The key outcome was overall survival (OS) of recipients in the preferred donor group. Specifically, we estimated the proportion of recipients alive at 60 months in the preferred donor group compared with the proportion alive for all recipients in the training set (reflecting standard of care). The results are shown in Fig. 2, with the average survival of the 10 cross-validation samples plotted. Classifier A, the most stringent, identified about 13% in the preferred donor group. 66.7% of patients in the preferred donor group per Classifier A were alive at 60 months, compared with the standard of care average of 44.9%. Classifier F, the least stringent, identified about 80% of patients in the preferred donor group. 46.3% of patients who received preferred donor stem cells were alive at 60 months. In our analysis, preferred donors were associated with lower relapse rates at 12 and 24 months, higher rates of remission at the time of death, and lower transplant-related mortality.
Next steps include validation of classifier performance using an independent set of 320 donor samples from the CIBMTR.
References:
1. Gadalla, S.M., et al., JAMA, 2015. 313(6): p. 594-602.
2. Krstajic, D., et al., Journal of Cheminformatics, 2014. 6(10).
Lee:Bristol-Myers Squibb: Consultancy; Kadmon: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.