Abstract

AML and MDS are heterogeneous myeloid neoplasms with variable biologic and clinical outcomes. Although allogeneic HCT is the only potentially curative therapy for high risk AML and MDS, survival after transplant remains poor, and identifying who benefits is challenging.
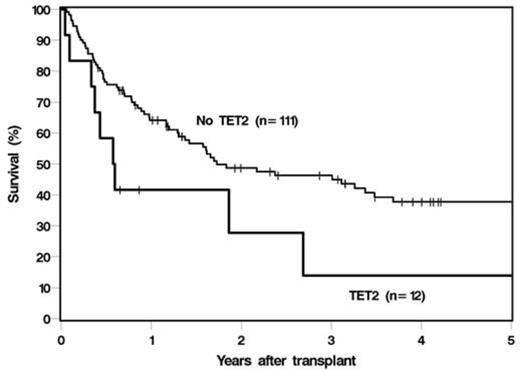
We hypothesized that next-generation sequencing (NGS) mutational analyses can predict outcome in MDS and AML patients undergoing allogeneic HCT. We performed multi-amplicon targeted pre-HCT NGS using a somatic panel of the 60 most commonly mutated genes in myeloid neoplasias as previously determined by whole exome sequencing, on 123 patients with AML (N=64, 52%) and MDS (N=59, 48%) who subsequently underwent HCT. Median age at transplant was 53 years (range, 20-73). 21 (17%) patients had complex karyotype, 10 (8%) with monosomy 7, 48 (39%) normal, and 48 (39%) with other or unknown cytogenetic abnormalities. 45 (37%) patients were in a complete remission (CR) prior to transplant, while 78 (63%) were in less than a CR; with CR as defined by International Working Group criteria for MDS, or <5% blasts for AML. The majority of patients received myeloablative conditioning (N=83, 68%), and 40 (33%) received a reduced-intensity preparative regimen. Donor source was matched sibling (N=52, 42%), matched unrelated (N=56, 46%), cord-blood (N=12, 10%), and haplo-identical (N=3, 2%). Median follow up was 35 months (range 5-178). Mutations were analyzed individually and by molecular pathway. 88 (72%) patients had at least one mutation, most frequently in STAG2 (10.2%), TET2 (9.8%), ASXL1 (8.1%), and RUNX1 (8.1%). TP53 mutations were more common in MDS patients compared to AML (10% versus 1.6%, P=0.05). NRAS (P=0.019) and TP53 (P=0.022)mutations were more commonly associated with complex karyotype. Mutations in BCOR (P=0.048) and TP53 (P=0.047)were associated with less than CR, while TET2 (P=0.03)mutations were associated with CR prior to HCT. In univariable analyses, the presence of complex karyotype was associated with shorter overall (OS) and relapse-free survival (RFS) (hazard ratio [HR] 2.4; P=0.002 and HR 3.1; P<0.001). Mutations in TET2 (HR 2.1; P=0.042) and EZH2 (HR 2.3; P=0.048), or presence of any mutation in the histone modification pathway (ASXL1, EZH2, KDM6A, SUZ12); (HR 1.7; P=0.039) was associated with poor OS. The presence of any mutation in the DEAD box RNA-helicase family genes (DHX29, DDX54, DDX41) was associated with poor RFS (HR 3.1; P=0.009). Nothing except complex karyotype was specifically associated with higher relapse. Unlike in previous reports, TP53 mutations were not found to be significantly associated with poor OS or RFS, though these cases (N=7) were limited. In multivariable analyses, adjusting for clinical variables, complex karyotype remained significantly associated with poor OS (HR 2.7; P<0.001) and RFS (HR 3.9; P<0.001). TET2 also remained independently associated with poor OS (HR 2.4; P=0.022). Presence of any of the DNA methylation mutations (TET2, DNMT3A, IDH1, IDH2) was associated with poor RFS (HR 1.7; P=0.05). 3-year OS was 23% in patients with a complex karyotype versus 48% in patients without (P=0.002); and 14% in patients with a TET2 mutation and 46% without (P=0.042) (Figure 1).
Molecular abnormalities are important variables in determining outcome after allogeneic HCT. We demonstrate that TET2 mutations in AML and MDS predict for poor survival after HCT. Ongoing serial mutational analyses in an extended cohort of patients will enhance our understanding of the role of NGS in informing care decisions for patients undergoing allogeneic HCT for AML and MDS.
Majhail:Gamida Cell Ltd.: Consultancy; Anthem Inc.: Consultancy. Sekeres:Celgene Corporation: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This icon denotes a clinically relevant abstract