Key Points
Mutations in HSPA9 cause CSAs that may be inherited in a recessive or pseudodominant manner.
HSPA9 loss-of-function alleles are often inherited in trans with a common coding single nucleotide polymorphism associated with altered gene expression.
Abstract
The congenital sideroblastic anemias (CSAs) are relatively uncommon diseases characterized by defects in mitochondrial heme synthesis, iron-sulfur (Fe-S) cluster biogenesis, or protein synthesis. Here we demonstrate that mutations in HSPA9, a mitochondrial HSP70 homolog located in the chromosome 5q deletion syndrome 5q33 critical deletion interval and involved in mitochondrial Fe-S biogenesis, result in CSA inherited as an autosomal recessive trait. In a fraction of patients with just 1 severe loss-of-function allele, expression of the clinical phenotype is associated with a common coding single nucleotide polymorphism in trans that correlates with reduced messenger RNA expression and results in a pseudodominant pattern of inheritance.
Introduction
The sideroblastic anemias are a heterogeneous group of inherited and acquired disorders characterized by the presence of ring sideroblasts (erythroblasts containing pathological mitochondrial iron deposits) in the bone marrow. Congenital sideroblastic anemias (CSAs) are associated with germline mutations leading to defects in mitochondrial heme synthesis, iron-sulfur (Fe-S) cluster biogenesis, or protein synthesis.1,2 In particular, mutations in GLRX5 and ABCB7, involved in mitochondrial Fe-S cluster biosynthesis and maturation of cytosolic Fe-S cluster proteins, cause nonsyndromic and syndromic forms of CSA, respectively.3-5 Here, we demonstrate that mutations in HSPA9, a mitochondrial HSP70 homolog involved in mitochondrial Fe-S biogenesis, result in an autosomal recessive CSA. The disorder has an unusual feature: a large fraction of patients carry a common coding single nucleotide polymorphism (cSNP) associated with altered messenger RNA (mRNA) expression in trans with a loss-of-function allele, resulting in pseudodominant inheritance in some families.
Study design
We performed genotyping for family linkage and copy number variation screening by using Affymetrix 6.0 chips. Analysis was performed by using the custom-built, rule-based algorithm Variant Explorer. We identified candidate genes likely to play a role in CSA based on pathway analysis and comparison with the MitoCarta database. Standard Sanger sequencing, quantitative reverse-transcription polymerase chain reaction, western blotting, and yeast genetic techniques were used to detect and further characterize sequence variants. For more information, see the supplemental Data available on the Blood Web site.
Results and discussion
We performed linkage analysis on a Dutch pedigree with mild CSA inherited in an apparently autosomal dominant manner (Table 1 and Figure 1A, family A)6 and mapped the disease to chromosome 5q (logarithm of odds [LOD], 3.00). A second family (family B) also demonstrated dominant linkage to 5q (LOD, 1.20; cumulative LOD, 4.20), defining a candidate interval of 26 Mb (chr5:134164092-160559870, HG19) and encompassing 241 genes. Because of the phenotype, we focused on the 14 mitochondrial proteins encoded therein.7 HSPA9, an HSP70 homolog, was the primary candidate because it is involved in mitochondrial Fe-S biogenesis,8-12 is highly expressed in erythroid precursors, is mutated in the zebrafish anemia crimsonless mutant,13 and is required for maturation of murine and human erythroid progenitors.14 Sequencing revealed a deletion of two nucleotides resulting in an early termination (NM_004134.6, c.409_410del, p.I137*) and an in-frame deletion of two amino acids (c.1373_1378del and p.del458_459) in affected individuals from families A and B, respectively (Figure 1B).
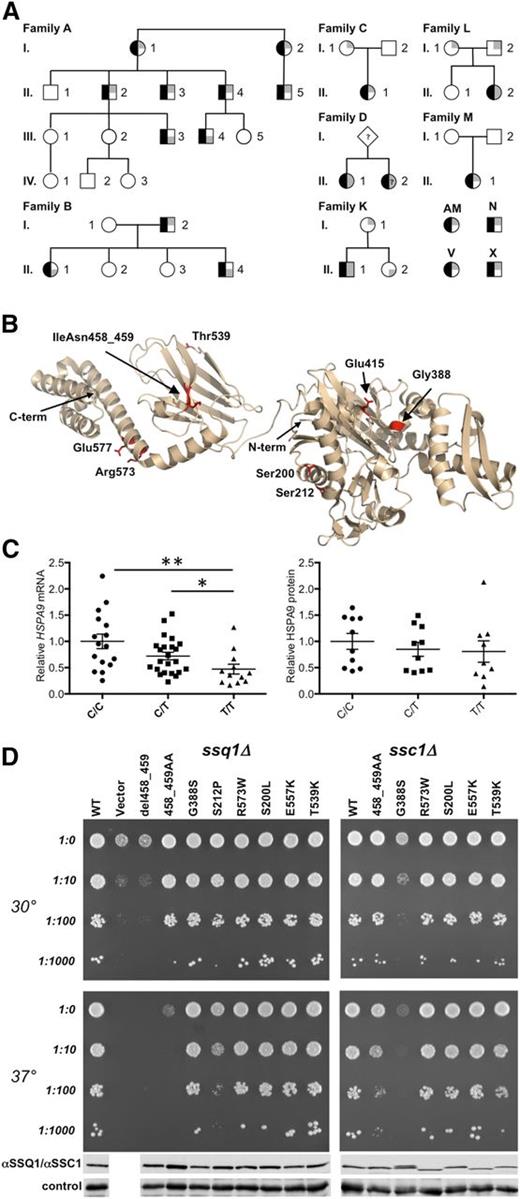
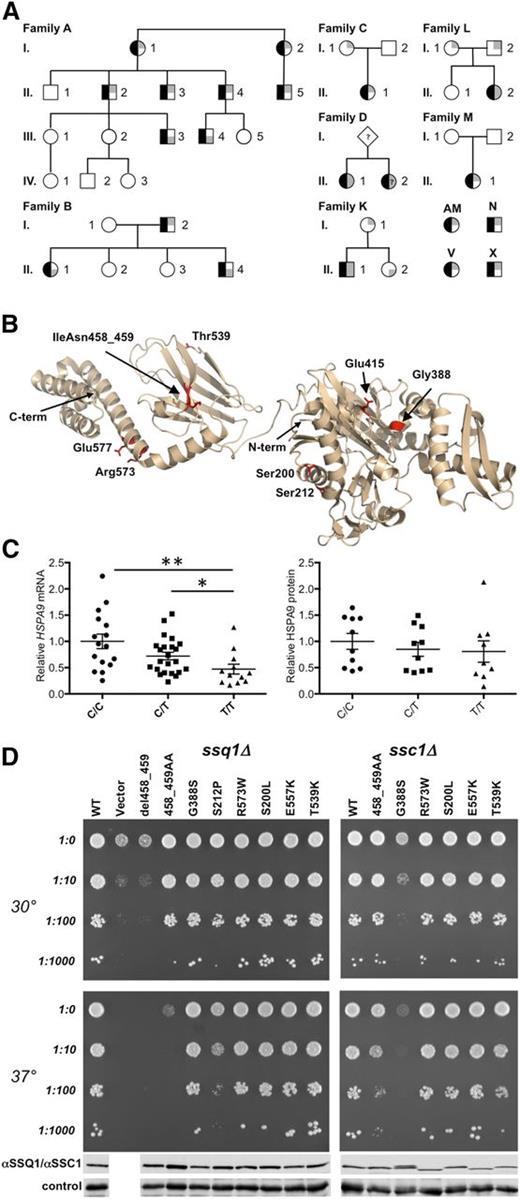
HSPA9 mutations in CSA. (A) HSPA9 CSA pedigrees. Affected status is indicated by black shading on the left of the symbol. The genotype is indicated on the right side of the symbol, in which gray shading in the upper and/or lower right quadrants indicates a low-frequency HSPA9 variant predicted to have functional consequences. When known, the paternal allele is indicated by shading in the lower right quadrant. A-I-1, A-I-2, A-II-1, A-II-2, A-II-3, A-II-4, A-II-5, A-III-1, A-III-2, A-III-3, A-III-4, and A-III-5 refer to I.1, I.2, II.1, II.2, II.6, II.7, II.19, III.2, III.3, III.4, III.9, and III.10, respectively, in van Waveren Hogervorst et al.6 A-IV-1, A-IV-2, and A-IV-3 were not included in van Waveren Hogervorst et al.6 Patient D-II-2 was not available for genotyping but had a phenotype identical to that of her sibling. (B) Mutations in HSPA9 mapped on the structure of bacterial HSP70 (Protein Data Bank ID: 2KHO). The N- and C-termini (term.) of the structure are noted. Human HSPA9 residues Ser200, Ser212, Gly388, Glu415, IleAsn458_459, Thr539, Arg573, and Glu577 were mapped to equivalent bacterial residues Ala149, Ala161, Gly342, Glu369, Ile412-Ala413, Ser493, Arg527, and Glu531. (C) Analysis of HSPA9 expression. Total mRNA was harvested from leukocytes (rs10117 genotype: number of samples analyzed; C/C: n = 16; C/T: n = 23; and T/T: n = 12); HSPA9 mRNA was assessed by quantitative real-time polymerase chain reaction and was normalized to β-actin. P values were calculated by using the Mann-Whitney test. **P < .005; *P < .05. Western blot analysis of HSPA9 protein expression in healthy individuals was grouped by rs10117 allele (C/C: n = 10; C/T: n = 10; and T/T: n = 9). Equivalent loading of mitochondrial lysate was confirmed by immunoblot analysis using an anti-adenosine triphosphate synthase, beta-subunit (ATPB) antibody. Protein expression was determined by densitometry analysis on a Biorad Chemidoc MP instrument with Image Laboratory 4.1 software. (D) Haploid ssq1Δ (left) and ssc1∆ (right) strains having plasmids harboring the indicated mutants, wild-type (WT) gene, or in the case of the viable ssq1∆ strain, a plasmid-lacking insert (vector). Tenfold serial dilutions were plated on minimal media and incubated at the indicated temperatures. Whole-cell lysates of the indicated strains were subjected to immunoblot analysis using polyclonal antibodies specific for SSQ1 (left), SSC1 (right) and, as a loading control, Ydj1. Names of the alleles correspond to the human HSPA9 numbering. For details of homologous yeast mutations, see supplemental Figure 1.
HSPA9 mutations in CSA. (A) HSPA9 CSA pedigrees. Affected status is indicated by black shading on the left of the symbol. The genotype is indicated on the right side of the symbol, in which gray shading in the upper and/or lower right quadrants indicates a low-frequency HSPA9 variant predicted to have functional consequences. When known, the paternal allele is indicated by shading in the lower right quadrant. A-I-1, A-I-2, A-II-1, A-II-2, A-II-3, A-II-4, A-II-5, A-III-1, A-III-2, A-III-3, A-III-4, and A-III-5 refer to I.1, I.2, II.1, II.2, II.6, II.7, II.19, III.2, III.3, III.4, III.9, and III.10, respectively, in van Waveren Hogervorst et al.6 A-IV-1, A-IV-2, and A-IV-3 were not included in van Waveren Hogervorst et al.6 Patient D-II-2 was not available for genotyping but had a phenotype identical to that of her sibling. (B) Mutations in HSPA9 mapped on the structure of bacterial HSP70 (Protein Data Bank ID: 2KHO). The N- and C-termini (term.) of the structure are noted. Human HSPA9 residues Ser200, Ser212, Gly388, Glu415, IleAsn458_459, Thr539, Arg573, and Glu577 were mapped to equivalent bacterial residues Ala149, Ala161, Gly342, Glu369, Ile412-Ala413, Ser493, Arg527, and Glu531. (C) Analysis of HSPA9 expression. Total mRNA was harvested from leukocytes (rs10117 genotype: number of samples analyzed; C/C: n = 16; C/T: n = 23; and T/T: n = 12); HSPA9 mRNA was assessed by quantitative real-time polymerase chain reaction and was normalized to β-actin. P values were calculated by using the Mann-Whitney test. **P < .005; *P < .05. Western blot analysis of HSPA9 protein expression in healthy individuals was grouped by rs10117 allele (C/C: n = 10; C/T: n = 10; and T/T: n = 9). Equivalent loading of mitochondrial lysate was confirmed by immunoblot analysis using an anti-adenosine triphosphate synthase, beta-subunit (ATPB) antibody. Protein expression was determined by densitometry analysis on a Biorad Chemidoc MP instrument with Image Laboratory 4.1 software. (D) Haploid ssq1Δ (left) and ssc1∆ (right) strains having plasmids harboring the indicated mutants, wild-type (WT) gene, or in the case of the viable ssq1∆ strain, a plasmid-lacking insert (vector). Tenfold serial dilutions were plated on minimal media and incubated at the indicated temperatures. Whole-cell lysates of the indicated strains were subjected to immunoblot analysis using polyclonal antibodies specific for SSQ1 (left), SSC1 (right) and, as a loading control, Ydj1. Names of the alleles correspond to the human HSPA9 numbering. For details of homologous yeast mutations, see supplemental Figure 1.
Sequencing 88 other genetically undefined CSA probands identified 9 additional individuals with at least 1 HSPA9 variant with an allele frequency of <0.01% in the Exome Variant database (http://evs.gs.washington.edu/EVS/). This mutation burden alone is sufficient to implicate HSPA9 mutations as causative: 9 of 88 probands carried frameshift, nonsense, or nonsynonymous mutations, whereas only 63 such variants were present in 6258 individuals catalogued by the Exome Variant Server (EVS; P < 1.1 × 10−6) and 1372 variants in 60 000 individuals (P < 4 × 10−4) sequenced by the Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org/). Furthermore, 5 of the additional 9 probands carried frameshift mutations, whereas no such mutation was present in EVS (P < 1 × 10−9), and just 26 heterozygotes were present in ExAC (P < 1 × 10−14). In 3 probands (D-II-1, K-II-1, and L-II-2; Table 1), we identified two novel sequence variants. In families K and L, the mutations were biallelic by segregation; whole-exome sequencing of family L did not identify any other potentially causative mutations. In each of the individuals with 2 mutated alleles, 1 was a predicted null (p.V296* or pC487Sfs*3) and the other was a missense or splicing variant (p.S200L, p.E577K, c.609+10A>G; Figure 1B). In the remaining 5 individuals (AM, M-II-1, C-II-1, V, and X), we identified only 1 rare variant (p.V296*) and four missense alleles (p.S212P, p.G388S, p.E415K, and p.R573W) (Figure 1B). p.E415K was a de novo variant in patient M-II-1, whereas in family C, p.R573W was also present in the patient’s unaffected mother (C-I-1). In each case, there was no family history of anemia. Thus, some families with HSPA9 variations appeared to demonstrate autosomal recessive inheritance.
Given these contradictory data, we considered the possibility that affected individuals with only 1 uncommon sequence variant also cosegregated a deletion or a common HSPA9 allele that resulted in lower mRNA and/or protein expression. None of these families or any of the 79 remaining probands in the unexplained CSA cohort had an exonic copy number loss determined by either Affymetrix 6.0 chip analysis or quantitative digital droplet polymerase chain reaction. Furthermore, complete Sanger sequencing of intronic regions in all probands by using multiple splice prediction tools (www.umd.be/HSF/) did not identify any intronic sequence variants predicted to alter expression.15 However, the minor (T) sequence variant of the synonymous cSNP rs10117 (c.1933C>T, p.L645 = ) was present in trans of the mutant allele in 9 of 10 of the affected individuals that could be unambiguously phased from the apparently dominant families and in all 5 of the affected individuals carrying only 1 coding variant in which the variant was de novo (M) or in which there was no family information (AM, N, X, and V; Table 1). In 3 families (C, K, and L), the rs10117C allele was present in trans of the predicted deleterious HSPA9 coding variant in an unaffected parent or sibling; in 2 other families, the coding variant was present in trans of rs10117T. Thus, of 21 individuals heterozygous for a presumptive HSPA9 coding mutation, 14 of 16 affected individuals from 8 different families had rs10117T in trans, whereas 2 of 5 unaffected individuals from 3 other families had the rs10117T allele in trans. By analyzing these 21 individuals, a phenotype permutation test yielded P < .07, suggesting that rs10117T or a linked variant determines expression of the HSPA9 CSA phenotype in patients with a single unambiguously deleterious allele.
Analysis of rs10117 with 3 mRNA mutation analysis algorithms (www.umd.be/HSF/)15 independently predicted the sequence (CUGAAG) overlapping rs10117 to be an exonic splice enhancer that is disrupted by the rs10117T variation (UUGAAG). To validate this prediction, we compared the expression of HSPA9 mRNA and HSPA9 protein in peripheral blood from individuals of different rs10117 genotypes (Figure 1C). Individuals who were homozygous for the rs10117T variant expressed approximately half as much HSPA9 mRNA and four-fifths as much HSPA9 protein as those homozygous for the rs10117C variant; heterozygotes had intermediate levels. Importantly, there was substantial overlap in the level of expression in the homozygous groups, with some C allele homozygotes expressing as little mRNA or protein as the majority of T allele homozygotes. This indicates that the T allele itself is not deterministic, either because of the influence of genetic background or because it is in linkage disequilibrium with another functionally significant polymorphism. Likewise, this may account for the observation that occasional individuals with an unequivocal HSPA9 mutation in trans with the rs10117C or rs10117T alleles are phenotypically affected or unaffected, respectively.
To assess the functional consequences of the HSPA9 coding variations, we evaluated the phenotype of yeast expressing the analogous mutations in the HSPA9 orthologs SSQ1 and SSC1 (Figure 1D and supplemental Figure 1). The phenotype of p.del458_459 was indistinguishable from a deletion. Missense variants p.S200L, p.T539K, p.R573W, and p.E577K demonstrated very weak or no phenotype, whereas two others, p.S212P and p.G388S, had baseline or temperature-sensitive phenotypes in one or both of the ssq1Δ and/or ssc1Δ assays. No variant appeared to have a dominant negative phenotype in diploid yeast (data not shown). The p.E415K variant was not analyzed, but this missense substitution is predicted to be functionally severe because it is in the active site. Although the yeast complementation assays are imperfect, given the partial functional redundancy of SSC1 and SSQ1, no patient had a combination of either severe variant (p.S212P, p.del458_459, p.E415K, and p.G388S) and a predicted null allele; these variants are all co-inherited with a milder missense allele, potential splicing variant, or the rs10117T variant.
Taken altogether, the genetic and functional data suggest that germline incomplete loss of HSPA9 function results in a CSA phenotype. Notably, HSPA9 is one of the genes in the 5′ minimal 5q myelodysplastic syndrome deletion interval.16 The 5q syndrome is characterized by a defect in erythroid maturation, but ring sideroblasts are uncommon. Thus, an HSPA9 sideroblastic phenotype may be epistatically suppressed by other differentiation abnormalities in susceptible individuals,17 a hypothesis that warrants further investigation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the families for their ongoing participation in this research, Matt Walter and Ben Ebert for helpful discussions regarding the manuscript, and Erwin Wiegerinck and Siem Klaver for their help in measuring, collecting, and processing samples from family A.
M.D.F., K.M., and K.S.-A. were supported by grant R01 DK087992 from the National Institutes of Health (NIH) National Institute of Diabetes and Digestive and Kidney Diseases. I.H., M.D.F., K.M., and A.S. were supported by grant R24 DK099808 from the NIH National Institute of Diabetes and Digestive and Kidney Diseases. E.A.C. and S.J.C. were supported by grant R01 GM GM27870 from the NIH National Institute of General Medical Sciences. A.M. was supported by the Llandough Haematology Development Fund, and S.S.B. was supported by grants from the US Department of Veterans Affairs and the Oklahoma Center for Advancement of Science and Technology.
Authorship
Contribution: M.D.F., K.M., and E.A.C. designed and supervised the study; M.C., K.R., B.L., I.H., D.M., A.S., M.L.L., P.J.L.M.S., P.E.N., C.M.N., M.H.D., M.M.H., A.M., S.S.B., D.W.S., M.C.T., H.G.W., and M.D.F. characterized patients and collected clinical data and patient samples; K.S.-A. was supervised by K.M., developed the informatics pipeline, and identified the locus by way of linkage studies; S.J.C. performed yeast phenotyping and was supervised by B.A.S.; P.J.S. performed functional and expression analysis in primary human cells; D.R.C., C.G., and F.R. performed mutation analysis; A.K.S. performed structure-function studies and created Figure 1B; and P.J.S., M.D.F., E.A.C., and K.M. prepared the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mark D. Fleming, Boston Children’s Hospital, 300 Longwood Ave, Bader 124.1, Boston, MA 02115; e-mail: mark.fleming@childrens.harvard.edu.
References
Author notes
K.S.-A., S.J.C., P.J.S., and D.R.C. contributed equally to this work.