Key Points
A preclinical model system was developed to define the molecular landscape dictating lymphoma resistance to immunotherapy.
This system revealed that Gal-1 significantly inhibits CD20 immunotherapy within the lymphoma microenvironment in vivo.
Abstract
Non-Hodgkin lymphoma (NHL) is the most commonly diagnosed hematologic cancer of adults in the United States, with the vast majority of NHLs deriving from malignant B lymphocytes that express cell surface CD20. CD20 immunotherapy (rituximab) is widely used to treat NHL, even though the initial effectiveness of rituximab varies widely among patients and typically wanes over time. The mechanisms through which lymphomas initially resist or gain resistance to immunotherapy are not well established. To address this, a preclinical mouse model system was developed to comprehensively identify lymphoma transcriptomic changes that confer resistance to CD20 immunotherapy. The generation of spontaneous primary and familial lymphomas revealed that sensitivity to CD20 immunotherapy was not regulated by differences in CD20 expression, prior exposure to CD20 immunotherapy, or serial in vivo passage. An unbiased forward exome screen of these primary lymphomas was used to validate the utility of this expansive lymphoma cohort, which revealed that increased lymphoma galectin-1 (Gal-1) expression strongly correlated with resistance to immunotherapy. Genetically induced lymphoma Gal-1 expression ablated antibody-dependent lymphoma phagocytosis in vitro and lymphoma sensitivity to CD20 immunotherapy in vivo. Human NHLs also express elevated Gal-1 compared with nonmalignant lymphocytes, demonstrating the ability of this preclinical model system to identify molecular targets that could be relevant to human therapy. This study therefore established a powerful preclinical model system that permits the comprehensive identification of the dynamic lymphoma molecular network that drives resistance to immunotherapy.
Introduction
Non-Hodgkin lymphoma (NHL) is the most commonly diagnosed hematologic cancer of adults in the United States, with close to 70 000 people diagnosed in 2013 alone.1 NHL encompasses a heterogeneous variety of malignancies, of which 80% to 90% derive from B lymphocytes that express cell surface CD20.2 The CD20 monoclonal antibody (mAb) rituximab was the first biologic approved for the treatment of indolent NHL.3 CD20 mAb’s are now used to treat a number of human cancers, but there is wide variation in therapeutic efficacy between individual patients and among different cancers.4 The initial effectiveness of rituximab varies widely among patients and typically wanes over time, despite sustained CD20 expression by malignant cells among the majority of relapsing patients.4,5 Although rituximab is widely used, the molecular mechanisms by which human NHLs gain resistance to CD20 immunotherapy are unknown.6 Select human and mouse Fcγ receptor (FcγR) polymorphisms represent one explanation for reduced CD20 mAb efficacy.6-10 Inadequate clearance of endogenous interleukin (IL) 10–producing regulatory B cells (B10 cells) by CD20 mAb also significantly impairs lymphoma clearance by inhibiting monocyte activation in mice,11 with B10 cells also identified in humans.12 Tumor IL-10 production also inhibits antitumor immunity,13 and most chronic lymphocytic leukemias (CLLs) retain the functional capacity for IL-10 expression.14 Decreased or absent CD20 expression also accounts for reduced CD20 mAb efficacy in select patients, whereas other mechanisms accounting for lymphoma-intrinsic resistance to CD20 immunotherapy are not well described.6 Because most NHL patients eventually die of their disease despite CD20 immunotherapy, understanding the molecular basis for lymphoma resistance to therapy could lead to alternative and more effective treatment strategies.
A limited number of studies have searched for the molecular basis of lymphoma resistance to CD20 immunotherapy.6-9,15 However, heterogeneity between lymphomas, the predominant use of CD20 mAb in combination with other drugs, the inherent difficulty in quantifying therapeutic efficacy, and variable disease staging among patients in combination with their intrinsic genetic diversity makes these studies a daunting task. Therefore, the current study examined the molecular mechanisms responsible for lymphoma resistance to CD20 immunotherapy in a homologous mouse preclinical model of lymphoma. In this model, CD20 mAb therapeutic efficacy can be measured precisely under conditions where the host genetic background and timing of lymphoma initiation are identical. An extensive cohort of spontaneous primary lymphomas isolated from Eµ-cMyc transgenic mice was isolated and transferred into wild-type inbred mice that were then treated with mouse anti-mouse CD20 mAb as a monotherapy.16,17 This preclinical model of lymphoma development and progression through serial passage indicated that select expression changes led to lymphoma resistance to CD20 immunotherapy. A forward exome screen for the unbiased identification of lymphoma-intrinsic alterations identified a conserved gene product that is expressed in some human lymphomas, validating this approach for identifying lymphoma gene products with the potential to confer immunotherapy resistance in vivo.
Methods
Mice
B6.Cg-Tg(IghMyc)22Bri/J (Eμ-cMyc transgenic) hemizygous mice were from the Jackson Laboratory (Bar Harbor, ME).18,19 Lymphoma recipient and control C57BL/6 mice were from the National Cancer Institute Frederick Laboratory (Frederick, MD). The Duke University Animal Care and Use Committee approved all studies.
Lymphoma immunotherapy
Spontaneous monoclonal lymphomas isolated from the lymph nodes of individual Eμ-cMyc transgenic mice were cultured in complete RPMI medium consisting of RPMI 1640 (Cellgro, Herndon, VA), 10% to 20% fetal bovine serum (Sigma-Aldrich), 100 U/mL penicillin (Cellgro), 100 μg/mL streptomycin (Cellgro), 2 mM l-glutamine (Cellgro), and 55 μM 2-mercaptoethanol (Invitrogen, Carlsbad, CA). Lymphomas were cultured for 5 to 7 days to remove nonmalignant passenger cells and tumor debris before freezing in aliquots as described,11,17 although it is possible that some tumor selection may have occurred during this brief culture period. Viable cells were quantified by trypan blue staining using a hemocytometer. Thawed lymphoma cells were cultured for 24 to 48 hours in complete medium prior to injection of 1 × 105 viable cells in 250 μL phosphate-buffered saline (PBS) under the dorsal skin of recipient wild-type mice. Although this route of administration is nonorthotopic, subdorsal administration permits tumor volume measurement in living mice. Mice were given purified CD20 mAb (MB20-11)20 or unreactive mouse control IgG2c mAb (250 μg in 250 μL PBS) through lateral tail veins 1 day after lymphoma transfers, with daily monitoring for lymphoma development and mortality for 60 days. The half-life of the CD20 mAb used in these studies (MB20-11) is 4.6 days in mice, with the depletion of endogenous peripheral B cells after a single administration of MB20-11 lasting for close to 60 days.21 Mice without lymphomas by day 60 survived indefinitely and did not display overt signs of disease. Mice exhibiting distress or with lymphoma volumes that exceeded 2.0 cm3 when measured as described17 with a calibrated micrometer were euthanized.
Lymphomas were considered sensitive to CD20 immunotherapy if mice that had received lymphomas survived significantly longer when treated with CD20 mAb relative to mice treated with control mAb. Lymphoma sensitivity to CD20 immunotherapy was quantified by calculating the increase in the area under the curve (AUC) from 60-day Kaplan-Meier survival plots as follows: % sensitivity = [(AUC for CD20 mAb-treated mice) − (AUC for control mAb)] / (AUC for control mAb) × 100. Lymphomas obtained from mice used to determine the sensitivity of primary lymphomas to immunotherapy were collected and referred to as secondary lymphomas and then evaluated for sensitivity to immunotherapy in the same manner as primary lymphoma cells. This process was repeated for several generations to create lymphoma families derived from distinct primary lymphomas.
Immunofluorescence analysis
CD20 expression was visualized using the MB20-11 mouse CD20 mAb16 conjugated using Pacific Blue Succinimidyl Ester (Invitrogen). Other mAb’s included the following: phycoerythrin (PE)-, allophycocyanin-, and phycoerythrin-cyanine 7 (PECy7)–conjugated CD19 (6D5), CD21/35 (7E9), B220 (RA3-6B2), streptavidin, and isotype controls from Biolegend (San Diego, CA); PE- and PECy7-conjugated CD5 (53-7.3), CD23 (B3B4), and IgM (II/41) from Ebioscience (San Diego, CA); PE-conjugated CD24 (M1/69) from BD Pharmingen (San Jose, CA); and biotinylated IgD (11-26C) from Southern Biotechnology Associates (Birmingham, AL).
Single-cell suspensions (1 × 106 per sample) were stained on ice for 30 minutes using optimal concentrations of mAb, Fc block (Clone 93, Biolegend), and LIVE/DEAD Fixable Red Dead Cell Stain Kit (Invitrogen) in PBS according to the manufacturer’s protocols.22 Cells were washed between all staining steps in fluorescence-activated cell sorter (FACS) buffer (2% fetal calf serum in PBS) and were resuspended in PBS containing 1.5% paraformaldehyde after staining. Immunofluorescence staining of single live cells was analyzed using a FACSCanto II flow cytometer (BD Biosciences), with background staining levels determined using isotype- and fluorochrome-matched control mAb’s. Control mAb mean linear fluorescence staining intensities (MFIs) were subtracted from each lymphoma MFI staining value.
Macrophage phagocytosis assay
Peritoneal macrophages were collected by lavage 4 days after intraperitoneal thioglycollate injection and were allowed to adhere to tissue culture dishes overnight. BL3750 lymphoma cells that expressed green fluorescent protein (GFP) were labeled with CellTrace Violet (Invitrogen) using the manufacturer’s protocols to track phagocytosed cells because GFP is rapidly degraded within macrophage lysosomes.23-25 Lymphomas stained with control isotype or MB20-11 mAb as described previously were added to the macrophage cultures at a 1:1 ratio (2 × 106 cells per mL total), and then cultures were incubated for 3, 6, or 24 hours. Recombinant galectin-1 (rGal-1; 5 ng/mL; R&D Systems, Minneapolis, MN) was added to some cultures as indicated. Control cocultures were incubated at 4°C for the duration of the experiment. Nonadherent cells were collected, then adherent cells were removed using trypsin-EDTA and were stained as described previously. Resident and thioglycollate-elicited peritoneal macrophages had comparable rates of antibody-dependent phagocytosis, consistent with their clearance of lymphomas in vivo through antibody-dependent mechanisms.11,17,26,27 Thioglycollate-elicited macrophages were used for assays in the present data to ensure sufficient cell counts for necessary controls in each assay.
Gal-1 studies
Total RNA was extracted from lymphoma cells using TRIzol (Invitrogen), with oligoDT primers used to generate complementary DNA (cDNA; Promega, Madison, WI). Relative transcript levels were quantified by GeneChip analysis (Affymetrix Mouse Genome 430 2.0 GeneChips; Affymetrix, Santa Clara, CA). All quality parameters for the arrays were confirmed to be in the manufacturer’s recommended range. Quantitative Lgals1 transcript levels were determined by reverse-transcription polymerase chain reaction amplification of triplicate samples using Bio-Rad reagents and an iCycler iQ system (Bio-Rad, Hercules, CA) as described using gapdh primers28 and Lgals1 primers (forward, CGCCAGCAACCTGAATC; reverse, GTCCCATCTTCCTTGGTGTTA). Reverse-transcription polymerase chain reaction product specificity was confirmed by melting curve analyses and sequencing. Transcript data are shown as Lgals1 (mouse) or LGALS1 (human) transcript levels relative to gapdh or internal housekeeping transcripts. To measure Gal-1 protein secretion, individual lymphomas (1 × 106 cells per mL) were cultured in triplicate for 24 hours prior to Mouse Galectin-1 DuoSet ELISA kit (R&D Systems) quantification of culture supernatant fluid.
BL3750Ctrl and BL3750Gal-1 lymphomas were generated by retroviral infection of primary cells with the pMX-IRES-GFP plasmid with or without mouse Lgals1 cDNA inserted.29 Briefly, Lgals1 cDNA was amplified from primary splenic CD19+ B-cell RNA and introduced to the pMX-IRES-GFP downstream of the 5′ long terminal repeat sequence using BamHI and NotI sites, leaving enhanced GFP downstream of the internal ribosome entry site. Phoenix-Eco cells (ATCC, Rockville, MD) were then transduced with either control GFP or Gal-1-GFP expression plasmids, and virus was collected 3 days later for retroviral infection of BL3750 cells. GFP+ BL3750 cells were sorted by FACS 4 times to ensure homogeneity and plasmid expression stability. Serum for Gal-1 measurements was collected from mice given BL3750Ctrl or BL3750Gal-1 cells at day 45 post-mAb treatment.
Statistical analysis
All data, including outliers, were included in the presented experiments. Data are shown as individual data points, indicating unique mice or lymphomas, or as means (± standard error of the mean [SEM]). Significant differences in mouse survival were calculated using the log-rank test; significant differences in sample means were determined using the appropriate unpaired 2-tailed Student t test. Correlations between lymphoma sensitivity and CD20, Lgals1, or Gal-1 expression were determined by calculating Spearman’s correlation coefficient (ρ), with statistically significant linear regressions indicated by regression lines where appropriate and P values calculated by a Gaussian approximation. Comparative statistical analysis and the generation of Kaplan-Meier cumulative survival plots were performed with Prism software (version 5; GraphPad Software, San Diego, CA). Human microarray data (GSE2350) were analyzed using the Kruskal-Wallis 1-way analysis of variance with Dunn’s multiple comparison test for post hoc multiple pairwise comparisons.
Results
Lymphoma responses to CD20 immunotherapy do not correlate with CD20 expression levels
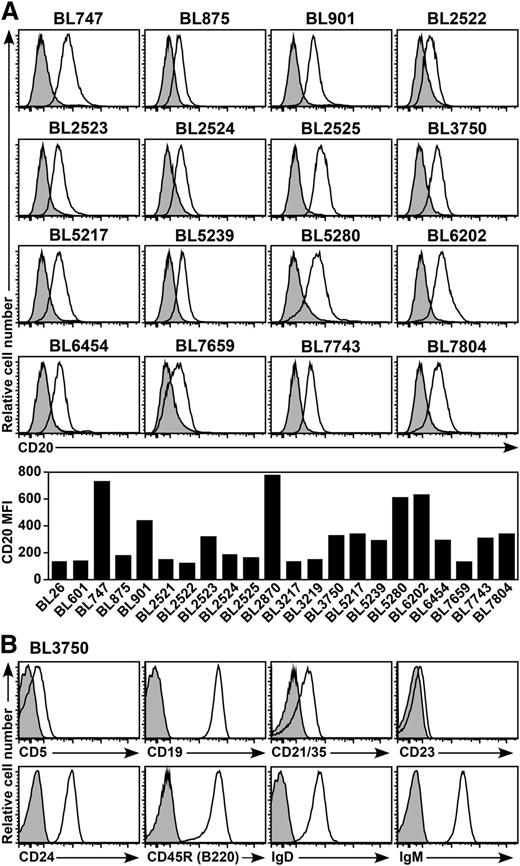
Twenty-two spontaneous B-cell lymphomas were isolated from Eμ-cMyc transgenic mice, which typically develop aggressive monoclonal Burkitt-like B-cell lymphomas by 6 months of age.18,19 Cell surface CD20 expression by each primary lymphoma was visualized by ex vivo immunofluorescence staining16 and varied between individual lymphomas (Figure 1A). All lymphomas generally shared a CD20+CD19+IgM+IgD+B220+CD5loCD21/35+CD23loCD24+ cell surface phenotype (Figure 1B), which is consistent with previous reports.17,30,31
CD20 expression by spontaneous mouse primary B-cell lymphomas. (A) Representative CD20 expression by primary B-cell lymphomas isolated from Eµ-cMyc transgenic mice. Cell surface CD20 (open histograms) and isotype-matched control (shaded histograms) immunofluorescence staining were quantified by flow cytometry after brief periods of culture for 24 to 48 hours. The bar graph shows CD20 MFIs with isotype-matched control values subtracted. (B) Representative cell surface phenotype of the BL3750 lymphoma. Histograms show cell surface molecule staining (open histograms) vs isotype-matched control mAb staining (shaded histograms).
CD20 expression by spontaneous mouse primary B-cell lymphomas. (A) Representative CD20 expression by primary B-cell lymphomas isolated from Eµ-cMyc transgenic mice. Cell surface CD20 (open histograms) and isotype-matched control (shaded histograms) immunofluorescence staining were quantified by flow cytometry after brief periods of culture for 24 to 48 hours. The bar graph shows CD20 MFIs with isotype-matched control values subtracted. (B) Representative cell surface phenotype of the BL3750 lymphoma. Histograms show cell surface molecule staining (open histograms) vs isotype-matched control mAb staining (shaded histograms).
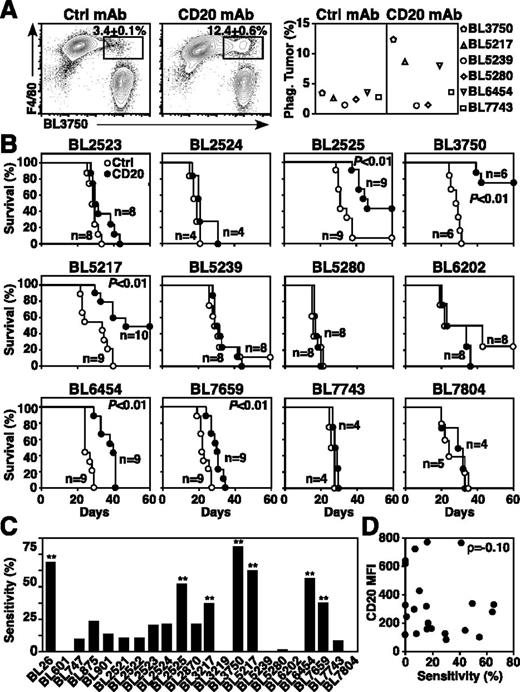
Normal and malignant B cells are depleted in mice following CD20 mAb treatment by macrophages through FcγR- and Ab-dependent cellular cytotoxicity/phagocytosis mechanisms independent of complement and cellular immunity.17,20,32 As macrophages are the predominant mediators of CD20 mAb-dependent B-cell depletion,17,20,32 primary lymphoma sensitivity to macrophage phagocytosis was evaluated. Macrophages were cultured for 24 hours with lymphomas that had been incubated with control or CD20 mAb. Although lymphoma survival in culture was equivalent, macrophages readily phagocytosed some lymphomas, such as BL3750, BL5217, and BL6454, whereas other lymphomas (BL5239, BL5280, BL7659) were resistant to CD20 mAb-dependent phagocytosis (Figure 2A).
Lymphoma sensitivity to CD20 mAb. (A) CD20 mAb-induced lymphoma phagocytosis by macrophages. Peritoneal macrophages were cocultured for 24 hours with dye-labeled primary lymphoma cells previously incubated with control or CD20 mAb. Representative contour plots of F4/80+ macrophages vs labeled BL3750 cells show mean (± SEM) frequencies of phagocytosed lymphoma cells within the indicated gates (left and middle panels). The graph shows mean results for the indicated representative lymphomas (n = 2 experiments per group). (B-D) CD20 mAb-induced lymphoma clearance in vivo. Mice given 1 × 105 primary lymphoma cells were treated with either CD20 (closed circles) or control (open circles) mAb 1 day later. (B) Kaplan-Meier survival plots of mice given the indicated lymphomas and CD20 or control mAb. Pooled results represent 2 to 3 independent experiments (n = 4-13 total mice per group) with significant cumulative survival differences between groups treated with CD20 vs control mAb indicated. (C) Primary lymphoma sensitivity to CD20 immunotherapy. Significant differences between sample means are indicated. **P ≤ .01. (D) CD20 expression (Figure 1A) and lymphoma sensitivity (B) to CD20 immunotherapy are not correlated.
Lymphoma sensitivity to CD20 mAb. (A) CD20 mAb-induced lymphoma phagocytosis by macrophages. Peritoneal macrophages were cocultured for 24 hours with dye-labeled primary lymphoma cells previously incubated with control or CD20 mAb. Representative contour plots of F4/80+ macrophages vs labeled BL3750 cells show mean (± SEM) frequencies of phagocytosed lymphoma cells within the indicated gates (left and middle panels). The graph shows mean results for the indicated representative lymphomas (n = 2 experiments per group). (B-D) CD20 mAb-induced lymphoma clearance in vivo. Mice given 1 × 105 primary lymphoma cells were treated with either CD20 (closed circles) or control (open circles) mAb 1 day later. (B) Kaplan-Meier survival plots of mice given the indicated lymphomas and CD20 or control mAb. Pooled results represent 2 to 3 independent experiments (n = 4-13 total mice per group) with significant cumulative survival differences between groups treated with CD20 vs control mAb indicated. (C) Primary lymphoma sensitivity to CD20 immunotherapy. Significant differences between sample means are indicated. **P ≤ .01. (D) CD20 expression (Figure 1A) and lymphoma sensitivity (B) to CD20 immunotherapy are not correlated.
The sensitivity of each lymphoma to CD20 immunotherapy in vivo was then measured by adoptively transferring lymphomas into syngeneic wild-type mice17 and treating mice 1 day later with either CD20 or control mAb (Figure 2B). CD20 mAb sensitivity was quantified as the increase in the area under the survival curves for CD20 vs control mAb-treated mice, ranging between 0% and ∼65% among primary lymphomas (Figure 2C). The BL26, BL2525, BL3217, BL3750, BL5217, BL6454, and BL7659 primary lymphomas were sensitive to CD20 immunotherapy (P ≤ .01), although most primary lymphomas (68%, n = 15 of 22) were CD20 mAb-resistant under these in vivo conditions. Notably, CD20 expression densities by primary lymphomas showed no correlation with sensitivity to CD20 immunotherapy (ρ = −0.10; Figure 2D).
CD20 immunotherapy does not select for treatment-resistant lymphomas
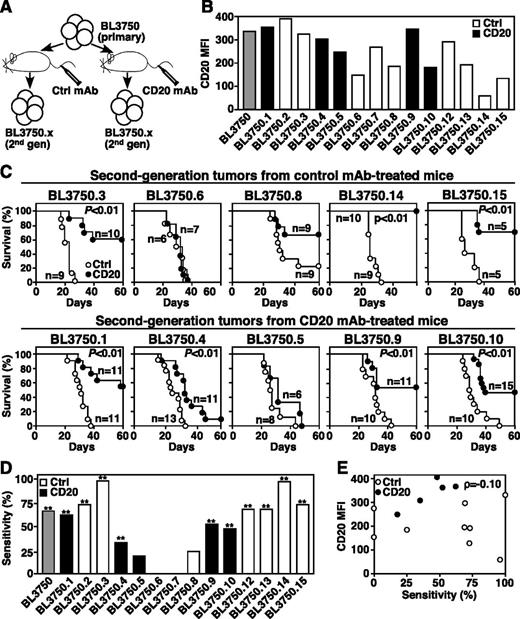
Following primary BL3750 lymphoma cell transfers and treatment with control or CD20 mAb (Figure 2), 14 lymphomas that developed in recipient syngeneic mice were collected and evaluated for CD20 expression and subsequent responsiveness to CD20 immunotherapy (Figure 3A). Secondary BL3750 lymphomas had varying CD20 expression densities, which did not correspond with prior exposure to CD20 or control mAb in vivo (Figure 3B). Most secondary lymphomas (71%, n = 10 of 14; P ≤ .01) were sensitive to CD20 immunotherapy, but prior exposure to CD20 or control mAb in vivo had no influence on lymphoma sensitivity to mAb treatment (Figure 3C-D). Similarly, CD20 expression by secondary lymphomas did not correlate with CD20 mAb sensitivity (ρ = −0.10; Figure 3E). Thus, CD20 immunotherapy does not drive immunoselection of lymphomas with reduced CD20 expression.
Second-generation lymphoma sensitivity to CD20 mAb. (A) Method for isolating second-generation BL3750 lymphomas from mice given primary cells. (B) CD20 expression by second-generation lymphomas with control mAb values subtracted. The gray bar indicates primary BL3750 cells; black bars indicate second-generation lymphomas from CD20 mAb-treated mice; white bars indicate second-generation lymphomas from control mAb-treated mice. (C) Second-generation lymphoma resistance to CD20 mAb is not driven by CD20 mAb selection in vivo. Mice were given second-generation lymphoma cells isolated from mice previously treated with either control (top panels) or CD20 (bottom panels) mAb’s as indicated. One day later, littermates were given either CD20 (closed circles) or control (open circles) mAb. Survival plots represent pooled results for 2 to 3 independent experiments (n = 5-15 mice per group) with significant differences between groups indicated. (D) Secondary lymphoma sensitivity to CD20 mAb as in Figure 2, with prior control or CD20 mAb exposure indicated. Significant differences between sample means are indicated. **P ≤ .01. (E) Second-generation lymphoma survival does not correlate with CD20 expression. Scatter plot comparing the percent sensitivity of secondary lymphoma cells with immunotherapy vs CD20 MFIs.
Second-generation lymphoma sensitivity to CD20 mAb. (A) Method for isolating second-generation BL3750 lymphomas from mice given primary cells. (B) CD20 expression by second-generation lymphomas with control mAb values subtracted. The gray bar indicates primary BL3750 cells; black bars indicate second-generation lymphomas from CD20 mAb-treated mice; white bars indicate second-generation lymphomas from control mAb-treated mice. (C) Second-generation lymphoma resistance to CD20 mAb is not driven by CD20 mAb selection in vivo. Mice were given second-generation lymphoma cells isolated from mice previously treated with either control (top panels) or CD20 (bottom panels) mAb’s as indicated. One day later, littermates were given either CD20 (closed circles) or control (open circles) mAb. Survival plots represent pooled results for 2 to 3 independent experiments (n = 5-15 mice per group) with significant differences between groups indicated. (D) Secondary lymphoma sensitivity to CD20 mAb as in Figure 2, with prior control or CD20 mAb exposure indicated. Significant differences between sample means are indicated. **P ≤ .01. (E) Second-generation lymphoma survival does not correlate with CD20 expression. Scatter plot comparing the percent sensitivity of secondary lymphoma cells with immunotherapy vs CD20 MFIs.
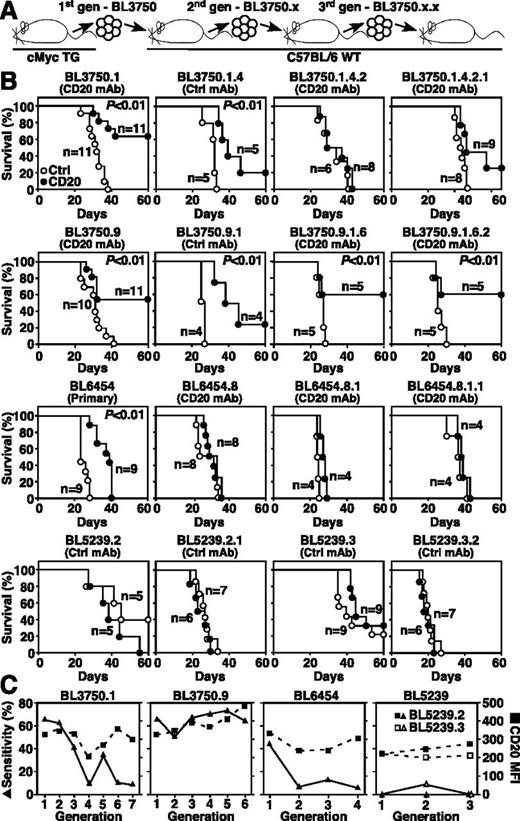
The effect of repeated in vivo passage and CD20 immunotherapy on lymphoma sensitivity to therapy was evaluated for lymphoma families derived from single primary lymphomas serially transferred over several generations (Figure 4A). In some instances, primary lymphomas that were initially sensitive to CD20 immunotherapy, such as BL3750.1 and BL6454, acquired resistance to CD20 immunotherapy during subsequent generations (Figure 4B). By contrast, other lymphomas from the same BL3750 family, such as BL3750.9, remained sensitive to CD20 immunotherapy over 6 generations. However, lymphomas that were initially resistant, such as BL5239, or lymphomas that became resistant to CD20 immunotherapy during repeated in vivo passages never reverted to a CD20 mAb-sensitive phenotype. Notably, changes in CD20 expression levels did not correlate with the development of resistance to CD20 immunotherapy (Figure 4C). Resistance to CD20 immunotherapy therefore appears to reflect lymphoma-intrinsic molecular changes that were independent of CD20 expression during in vivo progression.
CD20 immunotherapy does not select for treatment-resistant lymphomas. (A) Method for isolating and naming representative BL3750 lymphoma family members. Primary lymphoma cells were transferred into mice that were then given either CD20 (closed circles) or control (open circles) mAb as in Figures 2 and 3. Second-generation lymphomas were subsequently collected and adoptively transferred as in Figure 3. This process was repeated for subsequent generations. (B) Representative changes in BL3750, BL6454, and BL5239 lymphoma family member sensitivities to CD20 immunotherapy during progressive in vivo passages. Whether the transferred lymphomas were primary lymphomas or were isolated from mice treated with either control or CD20 mAb is indicated. Survival plots represent pooled results for 2 to 3 independent experiments (n = 4-11 total mice per group). Significant cumulative survival differences between groups are indicated. (C) CD20 immunotherapy during progressive in vivo passages does not select for CD20-deficient lymphomas. Line graphs compare the sensitivity of representative lymphoma cells with CD20 immunotherapy (closed triangles) vs their CD20 expression levels (closed squares) over 3 to 7 generations for the lymphoma families shown in panel B.
CD20 immunotherapy does not select for treatment-resistant lymphomas. (A) Method for isolating and naming representative BL3750 lymphoma family members. Primary lymphoma cells were transferred into mice that were then given either CD20 (closed circles) or control (open circles) mAb as in Figures 2 and 3. Second-generation lymphomas were subsequently collected and adoptively transferred as in Figure 3. This process was repeated for subsequent generations. (B) Representative changes in BL3750, BL6454, and BL5239 lymphoma family member sensitivities to CD20 immunotherapy during progressive in vivo passages. Whether the transferred lymphomas were primary lymphomas or were isolated from mice treated with either control or CD20 mAb is indicated. Survival plots represent pooled results for 2 to 3 independent experiments (n = 4-11 total mice per group). Significant cumulative survival differences between groups are indicated. (C) CD20 immunotherapy during progressive in vivo passages does not select for CD20-deficient lymphomas. Line graphs compare the sensitivity of representative lymphoma cells with CD20 immunotherapy (closed triangles) vs their CD20 expression levels (closed squares) over 3 to 7 generations for the lymphoma families shown in panel B.
Lymphoma families as a preclinical model of lymphoma development and progression
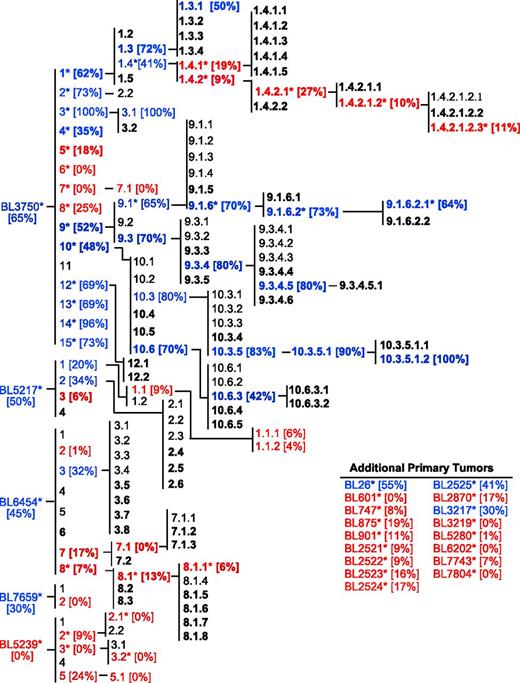
The correlation of lymphoma sensitivity to CD20 immunotherapy with in vivo progression was evaluated using the cohort of 22 primary lymphomas and their subsequent generations following in vivo passage, yielding a collection of 167 lymphomas comprising multiple families (Figure 5). Where tested, members of this lymphoma cohort were mostly resistant to CD20 immunotherapy (red text). CD20-negative lymphomas were never identified, and CD20 mAb resistance was unrelated to CD20 expression densities. Lymphomas that were either initially resistant or developed resistance to CD20 immunotherapy maintained treatment resistance through subsequent in vivo passages. By contrast, some lymphomas that were initially sensitive to CD20 immunotherapy (blue text) maintained sensitivity over time regardless of prior CD20 (bold text) or control mAb exposure in vivo (plain text). This extensive lymphoma family tree was therefore used for the unbiased identification of molecular alterations that contribute to CD20 mAb resistance during in vivo lymphoma progression.
Cumulative family tree for spontaneous primary lymphomas. Lymphoma families were evaluated over multiple generations for their sensitivity to CD20 mAb as in Figure 4. Lymphomas isolated from either CD20 mAb-treated mice (bold text) or control mAb-treated mice (plain text) were either resistant (red) or sensitive (blue) to CD20 immunotherapy. Bracketed numbers indicate lymphoma sensitivity to CD20 immunotherapy. A total of 167 lymphomas have been isolated, including the following: primary (n = 22), secondary (n = 34), tertiary (n = 45), quaternary (n = 38), quinary (n = 18), senary (n = 7), and septenary (n = 3) generations within families. Asterisks indicate specific lymphomas analyzed as in Figures 1-6 (n = 53).
Cumulative family tree for spontaneous primary lymphomas. Lymphoma families were evaluated over multiple generations for their sensitivity to CD20 mAb as in Figure 4. Lymphomas isolated from either CD20 mAb-treated mice (bold text) or control mAb-treated mice (plain text) were either resistant (red) or sensitive (blue) to CD20 immunotherapy. Bracketed numbers indicate lymphoma sensitivity to CD20 immunotherapy. A total of 167 lymphomas have been isolated, including the following: primary (n = 22), secondary (n = 34), tertiary (n = 45), quaternary (n = 38), quinary (n = 18), senary (n = 7), and septenary (n = 3) generations within families. Asterisks indicate specific lymphomas analyzed as in Figures 1-6 (n = 53).
Lymphoma Gal-1 expression confers resistance to CD20 immunotherapy
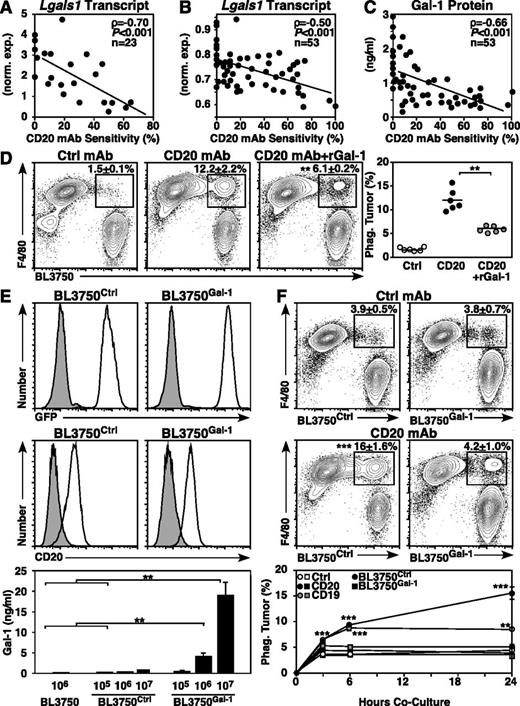
To identify molecular changes associated with CD20 immunotherapy resistance, transcript expression was evaluated in 10 primary, 7 BL3750 secondary, and 6 late-generation BL3750 family lymphomas. Although the expression of many genes was altered, one of the most significantly altered transcripts was Lgals1 (Gal-1), whereas other known immunosuppressive tumor-associated molecules,13 including the inhibitory cytokine IL-10,11,14,33 were unaltered. Although all lymphomas expressed Lgals1, Lgals1 expression levels correlated significantly with lymphoma sensitivity to CD20 immunotherapy (ρ = −0.70, P ≤ .001; Figure 6A). In a larger quantitative screen using 53 lymphomas (22 primary, 17 BL3750 secondary, and 14 late-generation lymphomas), Lgals1 transcript levels (ρ = −0.50, P ≤ .001; Figure 6B) and Gal-1 protein secretion (ρ = −0.66, P ≤ .001; Figure 6C) were significantly and inversely correlated with lymphoma sensitivity to CD20 immunotherapy. Notably, among all primary-generation lymphomas, Gal-1 secretion was even more closely associated with lymphoma sensitivity to CD20 immunotherapy (ρ = −0.77, P ≤ .001; data not shown).
Lymphoma Gal-1 expression correlates with CD20 mAb resistance. (A-C) Gal-1 expression inversely correlates with lymphoma sensitivity to CD20 immunotherapy. Scatter plots compare normalized (A) and quantitative (B) Lgals1 transcript expression and Gal-1 secretion (C) relative to CD20 mAb sensitivity for each lymphoma analyzed. (D) Addition of Gal-1 blocks CD20 mAb-dependent phagocytosis of lymphoma cells. Peritoneal macrophages were cocultured with labeled BL3750 lymphoma cells as in Figure 2A, with or without CD20 mAb or rGal-1 added to the cultures as indicated. Each dot represents the results of individual experiments, with bars indicating means. (E) Representative GFP and cell surface CD20 expression by BL3750Ctrl or BL3750Gal-1 cells (open histograms) vs BL3750 cells (top panels, shaded histograms) or isotype-matched control mAb staining (middle panels, shaded histograms). Values represent mean (± SEM) Gal-1 secretion by BL3750, BL3750Ctrl, and BL3750Gal-1 cells (bottom panel, pooled results from 5 experiments). (F) Gal-1 blocks CD20 mAb-dependent phagocytosis of lymphoma. Peritoneal macrophages were cocultured for 24 hours with labeled BL3750Ctrl or BL3750Gal-1 lymphomas previously incubated with control (top panels) or CD20 mAb (middle panels), with representative contour plots showing F4/80 vs labeled BL3750 staining and mean (± SEM) frequencies of phagocytosed lymphomas cells shown. Values represent mean (± SEM) frequencies of BL3750Ctrl (circles) or BL3750Gal-1 (squares) previously incubated with control (open shapes), CD20 (closed shapes), or CD19 mAb (gray shapes) and cultured for 3, 6, and 24 hours with macrophages from 3 to 5 experiments (bottom panel, n = 7-20 mice per group).
Lymphoma Gal-1 expression correlates with CD20 mAb resistance. (A-C) Gal-1 expression inversely correlates with lymphoma sensitivity to CD20 immunotherapy. Scatter plots compare normalized (A) and quantitative (B) Lgals1 transcript expression and Gal-1 secretion (C) relative to CD20 mAb sensitivity for each lymphoma analyzed. (D) Addition of Gal-1 blocks CD20 mAb-dependent phagocytosis of lymphoma cells. Peritoneal macrophages were cocultured with labeled BL3750 lymphoma cells as in Figure 2A, with or without CD20 mAb or rGal-1 added to the cultures as indicated. Each dot represents the results of individual experiments, with bars indicating means. (E) Representative GFP and cell surface CD20 expression by BL3750Ctrl or BL3750Gal-1 cells (open histograms) vs BL3750 cells (top panels, shaded histograms) or isotype-matched control mAb staining (middle panels, shaded histograms). Values represent mean (± SEM) Gal-1 secretion by BL3750, BL3750Ctrl, and BL3750Gal-1 cells (bottom panel, pooled results from 5 experiments). (F) Gal-1 blocks CD20 mAb-dependent phagocytosis of lymphoma. Peritoneal macrophages were cocultured for 24 hours with labeled BL3750Ctrl or BL3750Gal-1 lymphomas previously incubated with control (top panels) or CD20 mAb (middle panels), with representative contour plots showing F4/80 vs labeled BL3750 staining and mean (± SEM) frequencies of phagocytosed lymphomas cells shown. Values represent mean (± SEM) frequencies of BL3750Ctrl (circles) or BL3750Gal-1 (squares) previously incubated with control (open shapes), CD20 (closed shapes), or CD19 mAb (gray shapes) and cultured for 3, 6, and 24 hours with macrophages from 3 to 5 experiments (bottom panel, n = 7-20 mice per group).
The contribution of Gal-1 expression to CD20 mAb resistance was examined in mechanistic studies. To determine whether Gal-1 impairs mAb-dependent phagocytosis of lymphoma cells, macrophages were cultured for 24 hours with CD20 mAb-coated BL3750 cells with and without the addition of rGal-1. Lymphoma cells that were not exposed to mAb served as mAb-independent phagocytosis controls. The presence of soluble rGal-1 significantly reduced the phagocytosis of CD20 mAb-coated BL3750 cells (P ≤ .01; Figure 6D).
The impact of lymphoma-derived Gal-1 expression on CD20 mAb-dependent lymphoma clearance in vivo was then examined using parental BL3750 cells transfected with either Gal-1 GFP (BL3750Gal-1) or control GFP (BL3750Ctrl) expression plasmids. Genetically stable BL3750Gal-1 and BL3750Ctrl cells expressing comparable levels of GFP and cell surface CD20 were isolated (Figure 6E). BL3750Gal-1 cells secreted 10- to 20-fold more Gal-1 relative to BL3750Ctrl and untransfected BL3750 cells (P ≤ .01), and all cells had equivalent growth rates during in vitro cultures (data not shown). Importantly, BL3750Gal-1 cells and some CD20 immunotherapy-resistant primary lymphomas secreted Gal-1 at comparable levels (Figure 6C,E). To determine whether lymphoma-derived Gal-1 directly impedes mAb-dependent phagocytosis of lymphoma cells, macrophages were cultured with CD20 mAb-coated BL3750Ctrl or BL3750Gal-1 lymphoma cells for 3, 6, or 24 hours. Lymphomas that were not coated with mAb served as background phagocytosis controls. Macrophage survival was equivalent in all cultures with BL3750Ctrl and BL3750Gal-1 lymphomas. Macrophages readily phagocytosed CD20 mAb-coated BL3750Ctrl lymphomas, whereas lymphoma Gal-1 expression ablated CD20 mAb-dependent phagocytosis (P ≤ .001; Figure 6F). Gal-1 expression also inhibited macrophage phagocytosis of CD19 mAb-coated lymphomas (P ≤ .001), indicating that lymphoma Gal-1 expression broadly impedes mAb-dependent phagocytosis. FcγR expression by macrophages cultured with BL3750Gal-1 or BL3750Ctrl lymphomas was equivalent (data not shown), indicating that lymphoma-derived Gal-1 inhibited macrophage mAb-dependent phagocytosis through other mechanisms.
In vivo, Gal-1 expression by BL3750Gal-1 cells abrogated CD20 immunotherapy in mice (Figure 7A). By contrast, BL3750Ctrl cells remained sensitive to CD20 immunotherapy with 50% survival among mice given BL3750Ctrl cells and CD20 mAb (P ≤ .01). Importantly, BL3750Ctrl and BL3750Gal-1 lymphomas grew similarly in control mAb-treated littermates (median survival rates of 37 and 36.5 days, respectively). BL3750Gal-1 lymphomas also grew significantly larger than BL3750Ctrl lymphomas in CD20 mAb-treated mice, even though littermates bearing either BL3750Ctrl or BL3750Gal-1 lymphomas had similarly elevated serum Gal-1 (Figure 7B). Thus, lymphoma Gal-1 secretion within the tumor microenvironment inhibited CD20 immunotherapy.
Lymphoma Gal-1 expression in the local microenvironment confers CD20 mAb resistance in vivo. (A-B) Gal-1 expression blocks CD20-mediated lymphoma clearance in vivo. Mice given BL3750Ctrl or BL3750Gal-1 cells were treated with control (open shapes) or CD20 (closed shapes) mAb 1 day later. Lymphoma volume (2 experiments, top panels) and mouse survival (2-3 experiments, n = 4-10 mice per group, bottom panels) were monitored for 60 days post-mAb treatment. (B) Representative lymphomas (top panel) and serum Gal-1 (lower panel) was measured in naïve mice and littermates given lymphoma cells 45 days after CD20 mAb treatment (2-3 experiments, 10 mice per group). (C) Human LGALS1 expression (GSE2350) by naïve, centroblast (CB), centrocyte (CC), and memory B-cell samples (open circles) in comparison with cells (closed circles) from patients with Burkitt lymphoma (BL), CLL [blood cells (1) or blood CD19+ cells (2)], diffuse large B-cell lymphoma [DBCL; lymph node biopsy (1) or node biopsy CD19+ cells (2)], follicular lymphoma (FL), hairy cell leukemia (HCL), or mantle cell lymphoma (MCL). (E-I) Significant differences between sample means are indicated. *P ≤ .05; **P ≤ .01; ***P ≤ .001.
Lymphoma Gal-1 expression in the local microenvironment confers CD20 mAb resistance in vivo. (A-B) Gal-1 expression blocks CD20-mediated lymphoma clearance in vivo. Mice given BL3750Ctrl or BL3750Gal-1 cells were treated with control (open shapes) or CD20 (closed shapes) mAb 1 day later. Lymphoma volume (2 experiments, top panels) and mouse survival (2-3 experiments, n = 4-10 mice per group, bottom panels) were monitored for 60 days post-mAb treatment. (B) Representative lymphomas (top panel) and serum Gal-1 (lower panel) was measured in naïve mice and littermates given lymphoma cells 45 days after CD20 mAb treatment (2-3 experiments, 10 mice per group). (C) Human LGALS1 expression (GSE2350) by naïve, centroblast (CB), centrocyte (CC), and memory B-cell samples (open circles) in comparison with cells (closed circles) from patients with Burkitt lymphoma (BL), CLL [blood cells (1) or blood CD19+ cells (2)], diffuse large B-cell lymphoma [DBCL; lymph node biopsy (1) or node biopsy CD19+ cells (2)], follicular lymphoma (FL), hairy cell leukemia (HCL), or mantle cell lymphoma (MCL). (E-I) Significant differences between sample means are indicated. *P ≤ .05; **P ≤ .01; ***P ≤ .001.
Human lymphoma LGALS1 expression
LGALS1 transcript expression patterns among human B-cell leukemias and lymphomas was quantified using established data sets.34 Different B-cell subsets purified from nonreactive tonsils expressed relatively low levels of LGALS1 (Figure 7C), whereas biopsies or blood samples from patients with Burkitt lymphoma, CLL, diffuse large B-cell lymphoma, follicular lymphoma, hairy cell leukemia, and mantle cell lymphoma had on average 4.6-fold increased expression levels of LGALS1. Biopsies from patients with Burkitt lymphoma, diffuse large B-cell lymphoma, hairy cell leukemia, and mantle cell lymphoma expressed the highest levels of LGALS1 (average 6.3-fold, range 5.5- to 7.2-fold; P ≤ .001), whereas other tumor subtypes expressed modestly elevated LGALS1 transcript levels. Therefore, leukemia and lymphoma expression of Gal-1 may also inhibit CD20 immunotherapy in humans.
Discussion
The current study demonstrates the power of an unbiased forward exome screen to define the transcriptomic landscape dictating lymphoma resistance to immunotherapy using a preclinical model system. The spontaneous primary B-cell lymphomas examined in this study exhibited a wide spectrum of sensitivity to CD20 immunotherapy (Figures 1 and 2) that did not correlate with CD20 density (Figures 2 and 3) or time in vivo (Figures 3 and 4). Moreover, only a low threshold of lymphoma CD20 expression among primary and familial lymphomas was required for tumor clearance by CD20 mAb in vivo (Figures 2-4). Select primary lymphomas also acquired resistance to CD20 immunotherapy regardless of whether the lymphomas were exposed to CD20 or control mAb over multiple generations of passage (Figures 4 and 5). Thus, CD20 mAb resistance was primarily driven by the acquisition and evolution of alterations independent of CD20 immunoselection.
Lymphoma Gal-1 expression in the local microenvironment was identified as a significant mediator of resistance to CD20 immunotherapy and mAb-dependent phagoctyosis (Figures 6 and 7). All lymphomas expressed some level of Gal-1, which ranged from 0.2 to 3.0 ng/mL (Figure 6C). Although some resistant lymphomas secreted low amounts of Gal-1, all of the lymphomas that secreted >1 ng/mL Gal-1 were resistant to CD20 immunotherapy. Indeed, the 24-hour window between lymphoma transfer and CD20 mAb treatment was sufficient time for select lymphomas to secrete enough Gal-1 to in the local microenvironment to alter CD20 immunotherapy (Figure 6). Further, genetically induced Gal-1 overexpression in CD20 mAb-sensitive BL3750 cells, which secreted ∼4.0 ng/mL (Figure 6E), eliminated CD20 mAb-dependent clearance in vivo (Figure 7A). Mechanistically, exogenous rGal-1 as well as lymphoma-derived Gal-1 impaired mAb-dependent lymphoma phagocytosis by macrophages during short-term in vitro cultures (Figure 6D,F), demonstrating that extracellular Gal-1 impedes macrophage activation and/or function. However, not all CD20 mAb-resistant lymphomas secreted substantial Gal-1, indicating that these lymphomas must employ diverse mechanisms of resistance. The presence and potential combinations of these lymphoma-intrinsic molecular changes are likely to synergize and dictate therapeutic outcomes.
Galectins are evolutionarily conserved carbohydrate-binding proteins that modulate immune responses by impeding activation and enhancing cell death through cell-intrinsic and cell-extrinsic mechanisms.35 Gal-1 binds N-acetyllactosamine residues of N- and O-linked glycans on cell surface glycoproteins and impedes human and mouse macrophage, T-cell, and dendritic cell activation.36-41 Exposure to Gal-1 skews human and mouse dendritic cells to an anti-inflammatory phenotype and attenuates macrophage activation and FcγR and major histocompatibility complex class II induction as well as phagocytosis.40,42 Gal-1 secretion by melanoma cell lines inhibits T-cell–dependent tumor rejection.43 In the present study, Gal-1 did not alter macrophage survival or FcγR expression in vitro (data not shown) or mask CD20 mAb binding to cell surface CD20 (Figure 6E). It is unlikely that Gal-1 physically interferes with FcγR ligand binding because FcγR ligand affinity is multiple orders of magnitude greater than that of Gal-1 for its ligands (106-109 vs 105, respectively).44,45 Gal-1 most likely hinders macrophage activation and/or function directly, as has been previously observed,40,42 thereby impeding mAb-dependent phagocytosis of lymphoma cells in vivo. Indeed, lymphoma depletion relies on mAb-dependent macrophage function through FcγRs, so it is expected that Gal-1 expression would impair the function of other mAb-based therapies for human CLL and NHL, including those targeting CD20 (MabThera/rituximab, Roche; and Arzerra/ofatumumab, Genmab) and CD19.46,47 The efficacy of immunotherapies for solid tissue cancers, including receptor tyrosine-protein kinase erbB-2 (ERBB2) for breast and gastric cancers (Herceptin/trastuzumab, Genentech) and epidermal growth factor receptor for head and neck squamous cell carcinoma and colorectal cancers (Erbitux/cetuximab, Bristol-Myers Squibb), are also likely impeded by tumor Gal-1 production.48 It also remains possible that intracellular Gal-1 has profound impacts that additionally enhance lymphoma resistance to immunotherapy.49 Thus, Gal-1 expression within the lymphoma microenvironment has the potential to broadly impede diverse immunotherapies that rely on immune effector cell function.
Elevated serum Gal-1 levels correlate with disease severity in patients with CLL,50 B lymphoblastic leukemia,51 classic Hodgkin lymphoma,52,53 prostate tumors,54 and Kaposi sarcoma55 and were symptomatic of lymphoma burden in mice with BL3750Ctrl or BL3750Gal-1 lymphomas (Figure 7B). However, CD20 immunotherapy was effective in treating BL3750Ctrl but not BL3750Gal-1 lymphomas. Therefore, serum Gal-1, the source of which is unknown, may provide information regarding lymphoma burden but did not predict tumor resistance to immunotherapy in the present study. Human lymphoma Gal-1 expression may thereby provide a molecular signature for therapy-resistant lymphomas, and drugs that counteract the effects of Gal-1 expression within the local microenvironment may enhance tumor clearance during immunotherapy.
These studies developed an extensive preclinical model system that uses a comprehensive analysis of the exome to define the lymphoma-intrinsic regulatory networks that ultimately dictate lymphoma resistance to immunotherapy and potentially other drugs. Lymphoma resistance to CD20 immunotherapy is likely to be further complicated by inherent differences between patient immune responses as well as genetic and epigenetic changes in lymphomas. This reinforces the value of the current preclinical mouse model, where lymphoma-intrinsic alterations conferring resistance to single drug therapies can be identified in a comprehensive and unbiased fashion independent of the differential contributions of host genetic heterogeneity and environmental factors inherent to all patient populations. Further interrogation of the mouse lymphoma family tree outlined in this study will undoubtedly identify additional novel immunoregulators that contribute to therapy resistance and tumor immunologic privilege. Once identified and characterized in mice, these molecular changes and regulatory networks can be used to identify conserved therapeutic targets, to understand lymphoma-intrinsic regulatory networks, and to potentially forecast and enhance the biological outcome of immunotherapy in human cancer.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Marjolein van Egmond and Ayumi Yoshizaki for assistance with these studies.
This work supported by the Lymphoma Research Foundation, the National Institutes of Health, National Heart, Lung, and Blood Institute (training grant T32 HL007057) (J.M.L.), La Fondation de France (V.M.-C.), and La Federation Nationale des Centres de Lutte Contre le Cancer (V.M.-C.).
Authorship
Contribution: J.M.L., M.H., V.M.-C., M.K., T.M., J.C.P., and T.F.T. conceived of and designed the studies; J.M.L., M.H., V.M.-C., M.K., and T.M. carried out the experiments; and J.M.L., M.H., V.M.-C., J.C.P., and T.F.T. contributed to data analysis and manuscript preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas F. Tedder, Box 3010, Department of Immunology, Room 353 Jones Building, Research Dr, Duke University Medical Center, Durham, NC 27710; e-mail: thomas.tedder@duke.edu.
References
Author notes
J.M.L., M.H., and V.M.-C. contributed equally to this study.

![Figure 7. Lymphoma Gal-1 expression in the local microenvironment confers CD20 mAb resistance in vivo. (A-B) Gal-1 expression blocks CD20-mediated lymphoma clearance in vivo. Mice given BL3750Ctrl or BL3750Gal-1 cells were treated with control (open shapes) or CD20 (closed shapes) mAb 1 day later. Lymphoma volume (2 experiments, top panels) and mouse survival (2-3 experiments, n = 4-10 mice per group, bottom panels) were monitored for 60 days post-mAb treatment. (B) Representative lymphomas (top panel) and serum Gal-1 (lower panel) was measured in naïve mice and littermates given lymphoma cells 45 days after CD20 mAb treatment (2-3 experiments, 10 mice per group). (C) Human LGALS1 expression (GSE2350) by naïve, centroblast (CB), centrocyte (CC), and memory B-cell samples (open circles) in comparison with cells (closed circles) from patients with Burkitt lymphoma (BL), CLL [blood cells (1) or blood CD19+ cells (2)], diffuse large B-cell lymphoma [DBCL; lymph node biopsy (1) or node biopsy CD19+ cells (2)], follicular lymphoma (FL), hairy cell leukemia (HCL), or mantle cell lymphoma (MCL). (E-I) Significant differences between sample means are indicated. *P ≤ .05; **P ≤ .01; ***P ≤ .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/15/10.1182_blood-2015-11-681130/4/m_1886f7.jpeg?Expires=1767818824&Signature=ifJUi0LgApUNAOGV-vyX1uHO3Mhb0EXjK7OwUE7DjMeLaPI~jTvPdz9GsVJetVFHuOVbop~z0ATDTX4CZ2ca7CwPVkxneX63pbIW52eqqVdyTpOvAxfdwR609JY6-uw3JEJp2exW9Fj51v1DGKVDy7Ffu11Xbjdowr1AdFv7kkpX~hm59uLb0ajCHp4bcSdhkjLp6gQdYgJmnL64Pj6H8d8ahdTHlmGdhWrA5d9tqyojxjtNeN-13LJBD~uVL-FjCF5odByFKEhxdZ1BCBBZ0TSLjevVrq6wQVm4k24zSCeeIxz4ukrgT94w12qzSuiydJztuhSuN3O3WAERN04Itg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
