Key Points
Brca1 deficiency causes Fanconi anemia–like cytopenias, mitomycin C hypersensitivity, and spontaneous bone marrow failure.
Brca1 is critical for the maintenance of normal hematopoietic progenitor function and genomic stability in the bone marrow.
Abstract
BRCA1 is critical for maintenance of genomic stability and interacts directly with several proteins that regulate hematopoietic stem cell function and are part of the Fanconi anemia (FA) double-strand break DNA repair pathway. The effects of complete BRCA1 deficiency on bone marrow (BM) function are unknown. To test the hypothesis that Brca1 is essential in hematopoiesis, we developed a conditional mouse model with Mx1-Cre–mediated Brca1 deletion. Mice lacking Brca1 in the BM have baseline cytopenias and develop spontaneous bone marrow failure or diverse hematologic malignancies by 6 months of age. Brca1−/− BM cells have a reduced capacity to form hematopoietic colonies in vitro and to reconstitute hematopoiesis in irradiated recipients, consistent with a hematopoietic progenitor functional defect. Brca1−/− BM cells also show FA-like hypersensitivity to the DNA crosslinking agent mitomycin C, and karyotypes feature genomic instability. Taken together, our results show that loss of Brca1 in murine BM causes hematopoietic defects similar to those seen in people with FA, which provides strong evidence that Brca1 is critical for normal hematopoiesis and that Brca1 is a bona fide FA-like gene.
Introduction
Fanconi anemia (FA) is an inherited bone marrow failure (BMF) syndrome characterized by hypersensitivity to DNA crosslinking agents, congenital anomalies, BMF, and an increased risk of developing leukemia and solid tumors.1 FA is caused by mutations in one of 17 genes that make up the FA DNA repair pathway.2-4
BRCA1 binds directly to several FA proteins, all of which are essential for normal hematopoiesis, but the effects of BRCA1 deficiency on hematopoiesis are unknown. Several lines of evidence suggest that BRCA1 may also be an important regulator of hematopoiesis. First, BRCA1 is highly expressed in hematopoietic tissues,5,6 whereas its expression is lost in myeloid leukemias.7,8 Second, overexpression of Brca1 in the bone marrow (BM) disrupts stem cell quiescence and differentiation.9 Finally, the first person ever reported to have biallelic BRCA1 mutations experienced unusually severe myelosuppression after exposure to the DNA crosslinking agent carboplatin,10 which provides direct evidence that BRCA1 is important in human hematopoiesis.
To test whether BRCA1 has a critical role in hematopoiesis, we created a conditional Mx1-Cre–induced Brca1 deficiency mouse model. We demonstrated that mice with homozygous deficiency of Brca1 in the BM have FA-like cytopenias, DNA crosslinking agent hypersensitivity, and susceptibility to BMF and hematologic malignancies (HMs). Furthermore, Brca1−/− cells have a hematopoietic precursor functional defect characterized by a reduced capacity to form hematopoietic colonies in vitro and to reconstitute hematopoiesis in irradiated recipient mice, supporting a critical role for Brca1 in normal BM function.
Study design
Generation and monitoring of conditional Brca1-deficient mice
All mice were housed in a pathogen-free barrier facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care and maintained under a protocol approved by the Institutional Animal Care and Use Committee. Mice with a floxed Brca1 allele11 were bred with Mx1-Cre+ mice12 to generate Brca1+/+, Brca1+/−, and Brca1−/− mice after injections of polyinosinic:polycytidylic acid (supplemental Figure 1A-C available on the Blood Web site). Targeted Brca1 deletion was confirmed by polymerase chain reaction and real-time polymerase chain reaction (supplemental Figure 1C-D). Mice were monitored daily for the development of tumors, and they had monthly tail vein bleeds to measure complete blood counts. Full necropsy was performed on any mouse with changes in appearance that indicated illness.13 Histopathology, flow cytometry, and spectral karyotyping were performed on isolated tissues. Detailed methods are included in the supplemental Data.
Results and discussion
As early as 1 month of age, Brca1−/− mice demonstrated macrocytic anemia (hemoglobin [Hb] levels, 10.3 ± 3.3 vs 12.5 ± 3 g/dL; P = .042 [Figure 1A]; mean corpuscular volume 66.3 ± 5.0 vs 55.4 ± 4 fL; P < 1 × 10−5 [Figure 1B]) and had lower mean total white blood cell counts compared with Brca1+/+ mice (supplemental Figure 2).
Brca1 deficiency causes PB cytopenias, BM failure featuring genomic instability, and DNA crosslinking agent hypersensitivity. Complete blood counts from Brca1+/+ (red), Brca1+/− (green), and Brca1−/− (blue) mice were measured once per month up to 6 months of age. (A) Hb concentration (g/dL); (B) mean corpuscular volume (MCV) (fL). The number of mice in each cohort at each time point analyzed is listed in supplemental Figure 2. Analysis of variance was used to analyze for differences in counts at each time point (*P < .01). PB smears from (C) a Brca1+/+ (7145) and (D) a Brca1−/− (7386) mouse are shown (magnification ×10). BM sections from (E) a Brca1+/+ (7145) and (F) a Brca1−/− (7386) mouse are shown (magnification ×10). (G-H) Spectral karyotyping analysis revealed multiple structural abnormalities, including chromatid exchanges and premature centromere divisions. Representative cell karyotype in G: 40,XX,chte(2;5)(F1;C2),chte(9;12)(F1;E),pcd(16)(A)[1]. (I) Representative fluorescence-activated cell sorter plots from spleen cells from a Brca1+/+ (B7292) and a Brca1−/− (B7386) mouse stained with antibodies against CD71 and Ter119. (J) Average proportion of spleen cells accumulating in regions I, II, III, and IV of red blood cell differentiation after staining with antibodies against CD71 and Ter119. Student t test was used to analyze the differences in the proportion of cells within each region. (K) Sensitivity of Brca1−/− cells (blue) relative to Brca1+/+ cells (red) in methylcellulose colony-forming assays to mitomycin C at doses of 0, 5, 10, 25, and 50 nM. Averages are shown with standard error of the mean. chrb, chromosome break; chrg, chromosome gap; chtb, chromatid break; chte, chromatid exchange; chtg, chromatid gap; pcd, premature centromere division.
Brca1 deficiency causes PB cytopenias, BM failure featuring genomic instability, and DNA crosslinking agent hypersensitivity. Complete blood counts from Brca1+/+ (red), Brca1+/− (green), and Brca1−/− (blue) mice were measured once per month up to 6 months of age. (A) Hb concentration (g/dL); (B) mean corpuscular volume (MCV) (fL). The number of mice in each cohort at each time point analyzed is listed in supplemental Figure 2. Analysis of variance was used to analyze for differences in counts at each time point (*P < .01). PB smears from (C) a Brca1+/+ (7145) and (D) a Brca1−/− (7386) mouse are shown (magnification ×10). BM sections from (E) a Brca1+/+ (7145) and (F) a Brca1−/− (7386) mouse are shown (magnification ×10). (G-H) Spectral karyotyping analysis revealed multiple structural abnormalities, including chromatid exchanges and premature centromere divisions. Representative cell karyotype in G: 40,XX,chte(2;5)(F1;C2),chte(9;12)(F1;E),pcd(16)(A)[1]. (I) Representative fluorescence-activated cell sorter plots from spleen cells from a Brca1+/+ (B7292) and a Brca1−/− (B7386) mouse stained with antibodies against CD71 and Ter119. (J) Average proportion of spleen cells accumulating in regions I, II, III, and IV of red blood cell differentiation after staining with antibodies against CD71 and Ter119. Student t test was used to analyze the differences in the proportion of cells within each region. (K) Sensitivity of Brca1−/− cells (blue) relative to Brca1+/+ cells (red) in methylcellulose colony-forming assays to mitomycin C at doses of 0, 5, 10, 25, and 50 nM. Averages are shown with standard error of the mean. chrb, chromosome break; chrg, chromosome gap; chtb, chromatid break; chte, chromatid exchange; chtg, chromatid gap; pcd, premature centromere division.
Four (30%) of 13 Brca1−/− mice developed a steady decline in their Hb levels, eventually causing lethargy and resulting in euthanasia (supplemental Figure 3A). Necropsy confirmed severe anemia (mean Hb, 6.95 ± 0.6 g/dL), and peripheral blood (PB) smears showed polychromasia with irregular red blood cell morphology that was not present in Brca1+/+ mice (Figure 1C-D). BM sections were also hypocellular compared with age-matched Brca1+/+ mice (Figure 1E-F and supplemental Figures 8 and 9), consistent with spontaneous BMF. Spectral karyotyping of whole BM from 2 BMF Brca1−/− mice (Figure 1G-H and supplemental Figure 3B-C) revealed multiple cytogenetic abnormalities, resulting from 18 and 16 double-strand breaks detected in the 20 cells examined in each mouse, indicative of genomic instability.
By days 140 to 190, 6 additional Brca1−/− mice (6 [50%] of 13) developed HMs, including 4 lymphomas, 1 acute myeloid leukemia, and one erythroleukemia (Figure 2A-B).14,15 Trp53 sequencing identified DNA binding domain mutations in 2 of the lymphomas (Figure 2B). Histologic review of tissues from these mice demonstrated malignant cells circulating in the PB, disrupting hematopoietic organ architecture, and infiltrating nonhematopoietic organs (Figure 2C-J and supplemental Figure 9). Splenic cells isolated from the mouse with erythroleukemia were transplanted into 2 sublethally irradiated recipient mice, both of which developed erythroleukemia. Spectral karyotyping of malignant cells from the secondary transplant revealed multiple chromosomal translocations, a result of double-strand breaks (Figure 2K-L), indicative of genomic instability in a malignancy in this model as well. None of the Brca1+/+ or Brca1+/− mice developed a malignancy during 12 months of follow-up. These results demonstrate that lack of Brca1 within the BM leads to PB cytopenias, macrocytosis, spontaneous BMF, and susceptibility to HMs reminiscent of the hematopoietic defects observed in human FA.16,17
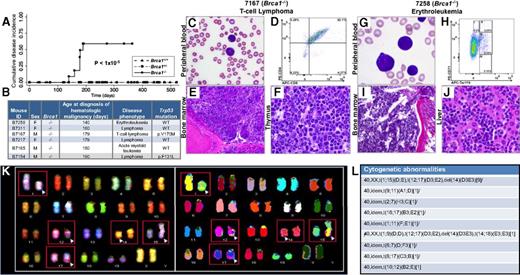
Brca1 deficiency increases susceptibility of mice to HMs characterized by leukemic infiltration of multiple organs consistent with the Bethesda criteria.14,15 (A) Cumulative disease incidence curves for Brca1+/+ (dash-dot-dot line), Brca1+/− (dashed line), and Brca1−/− (solid line) mice. Statistical significance was calculated by using the log-rank test. The number of mice in each cohort at each time point analyzed is listed in supplemental Figure 9H. (B) Characteristics of diseased mice and specific HM diagnosis. Presence or absence of a Trp53 mutation in each tumor is indicated. (C-F) Organs isolated from 7167, a Brca1−/−mouse that developed a T-cell lymphoma. (C) PB smear with Wright-Giemsa stain showing the presence of lymphoma cells (magnification ×10). (D) PB flow cytometry analysis using antibodies against T-lymphoid markers CD4 and CD8, showing the malignant cells to be CD4+/CD8+. (E) BM stained with hematoxylin and eosin (H&E), showing extensive involvement with lymphoma (magnification ×10). (F) Thymus stained with H&E, showing effacement of the normal thymic architecture by infiltrating lymphoma cells (magnification ×50). (G-J) Organs isolated from B7258, a Brca1−/− mouse that developed an erythroleukemia. (G) PB smear with Wright-Giemsa stain showing many erythroid blasts (magnification ×50). (H) PB flow cytometry analysis using antibodies against CD71 and Ter119. (I) BM stained with H&E, showing extensive leukemic involvement (magnification ×10). (J) Liver stained with H&E, showing extensive infiltration by leukemic cells (magnification ×50). (K) Spectral karyotyping analysis of erythroleukemia cells from a secondary transplant recipient mouse revealed an abnormal clone characterized by structural rearrangements: karyotype: 40,XX,t(1;15)(D;E), t(12;17)(D3;E2), and del(14)(D3E3). (L) Cytogenetic abnormalities identified within the erythroleukemia. del, deletion; idem, the same as the stemline clone listed first.
Brca1 deficiency increases susceptibility of mice to HMs characterized by leukemic infiltration of multiple organs consistent with the Bethesda criteria.14,15 (A) Cumulative disease incidence curves for Brca1+/+ (dash-dot-dot line), Brca1+/− (dashed line), and Brca1−/− (solid line) mice. Statistical significance was calculated by using the log-rank test. The number of mice in each cohort at each time point analyzed is listed in supplemental Figure 9H. (B) Characteristics of diseased mice and specific HM diagnosis. Presence or absence of a Trp53 mutation in each tumor is indicated. (C-F) Organs isolated from 7167, a Brca1−/−mouse that developed a T-cell lymphoma. (C) PB smear with Wright-Giemsa stain showing the presence of lymphoma cells (magnification ×10). (D) PB flow cytometry analysis using antibodies against T-lymphoid markers CD4 and CD8, showing the malignant cells to be CD4+/CD8+. (E) BM stained with hematoxylin and eosin (H&E), showing extensive involvement with lymphoma (magnification ×10). (F) Thymus stained with H&E, showing effacement of the normal thymic architecture by infiltrating lymphoma cells (magnification ×50). (G-J) Organs isolated from B7258, a Brca1−/− mouse that developed an erythroleukemia. (G) PB smear with Wright-Giemsa stain showing many erythroid blasts (magnification ×50). (H) PB flow cytometry analysis using antibodies against CD71 and Ter119. (I) BM stained with H&E, showing extensive leukemic involvement (magnification ×10). (J) Liver stained with H&E, showing extensive infiltration by leukemic cells (magnification ×50). (K) Spectral karyotyping analysis of erythroleukemia cells from a secondary transplant recipient mouse revealed an abnormal clone characterized by structural rearrangements: karyotype: 40,XX,t(1;15)(D;E), t(12;17)(D3;E2), and del(14)(D3E3). (L) Cytogenetic abnormalities identified within the erythroleukemia. del, deletion; idem, the same as the stemline clone listed first.
To investigate the etiology of these hematopoietic defects, we performed flow cytometry on single-cell suspensions from BM and spleen to assess for differences in specific hematopoietic cell populations. In the spleens of BMF mice, we found an accumulation of early red blood cell precursors at the basophilic erythroblast (CD71+, Ter119+) stage relative to Brca1+/+ mice (P = .003) (Figure 1I-J) that was not present in their BM (supplemental Figure 3D), suggesting a niche effect. We then assessed for differences in cell populations in the BM of 2-month-old Brca1−/− mice before the expected age of BMF or HM development vs age-matched Brca1+/− or Brca1+/+ mice. These experiments demonstrated an expansion of Lineage− Sca-1+ c-Kit+ cells, a population enriched for hematopoietic stem and progenitor cells (HSPCs) (supplemental Figure 5), but no differences in the numbers of long-term hematopoietic stem cells, short-term hematopoietic stem cells, multipotent progenitors, or other committed progenitors (supplemental Figures 5-7).
Because most other single-gene FA mouse models demonstrated reduced HSPC numbers and function,18,19 we tested the hematopoietic progenitor function of Brca1−/− BM cells. By using methylcellulose colony-forming assays, we found that Brca1−/− BM cells formed fewer hematopoietic colonies in vitro (44.4 ± 31.9) than Brca1+/+ cells (200.3 ± 30.5; P = .004). Similarly, BM cells from 2-month-old Brca1−/− mice were unable to reconstitute hematopoiesis in lethally irradiated congenic mice (supplemental Figure 4). These data demonstrate that although Lineage− Sca-1+ c-Kit+ cell numbers are expanded, HSPCs in our model have decreased function as seen in other FA models.
Finally, to test Brca1−/− BM cells for hypersensitivity to DNA crosslinking agents, a classic hallmark of FA, we performed colony-forming assays in the presence of increasing concentrations of mitomycin C. Brca1−/− BM cells demonstrated increased sensitivity compared with Brca1+/+ cells at all mitomycin C concentrations tested (Figure 1K). Taken together, these data demonstrate that Brca1 is critical for normal hematopoiesis and maintenance of genomic stability in the BM, and its deficiency results in an FA-like hematopoietic phenotype in mice.
Until recently, BRCA1 was not considered a true member of the FA pathway because the effects of homozygous deficiency of this gene in people were not known.3 Recently, 2 patients with biallelic BRCA1 mutations, each with 1 hypomorphic and 1 null allele, were reported. The FA-like phenotype in these patients included developmental anomalies and increased sensitivity of lymphocytes to chromosomal breakage with exposure to diepoxybutane,4,10 leading to the designation of BRCA1 as the seventeenth FA gene, FANCS. However, neither of these individuals has the characteristic FA BMF phenotype, leaving the effects of BRCA1 deficiency on BM function unknown.
Our data provide direct evidence of an FA-like hematopoietic defect in Brca1-deficient BM, strengthening the evidence that BRCA1 is a bona fide FA-like gene with a critical role in hematopoietic function. Furthermore, these mice provide a model system in which to determine the effects of Brca1 haploinsufficiency, as seen in humans, on BM function as well as ready access to a renewable source of Brca1-deficient tissue for characterization of the complex functions of Brca1. Finally, because nearly all other single FA gene mouse models have not recapitulated the BM phenotype of FA,18,20 further investigation of our model may provide novel insights into FA pathogenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Amittha Wickrema and Ryan Duggan for their helpful suggestions.
This work was funded by National Institutes of Health grants No. P01 CA40046 (M.M.L.B.) and K12CA139160 (J.E.C.) from the National Cancer Institute, K08HL129088 (J.E.C.) from the National Heart, Lung, and Blood Institute, and by the Cancer Research Foundation (M.M.L.B., L.A.G., J.E.C.) and Cancer Center Support grant No. P30 CA14599 from The University of Chicago Medicine Comprehensive Cancer Center.
Authorship
Contribution: A.V., M.M.L.B., L.A.G., and J.E.C. designed the research; A.V., S.A., R.M., J.L., G.R., A.A., M.W., R.D., and E.M.D. performed the research; A.V., S.A., R.M., G.R., B.N., E.M.D., M.M.L.B., J.W.V., L.A.G., and J.E.C. analyzed data; A.V. and J.E.C. wrote the manuscript; and all authors edited and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jane E. Churpek, The University of Chicago, 5841 S. Maryland Ave, MC2115, Chicago, IL 60637; e-mail: jchurpek@bsd.uchicago.edu.

![Figure 1. Brca1 deficiency causes PB cytopenias, BM failure featuring genomic instability, and DNA crosslinking agent hypersensitivity. Complete blood counts from Brca1+/+ (red), Brca1+/− (green), and Brca1−/− (blue) mice were measured once per month up to 6 months of age. (A) Hb concentration (g/dL); (B) mean corpuscular volume (MCV) (fL). The number of mice in each cohort at each time point analyzed is listed in supplemental Figure 2. Analysis of variance was used to analyze for differences in counts at each time point (*P < .01). PB smears from (C) a Brca1+/+ (7145) and (D) a Brca1−/− (7386) mouse are shown (magnification ×10). BM sections from (E) a Brca1+/+ (7145) and (F) a Brca1−/− (7386) mouse are shown (magnification ×10). (G-H) Spectral karyotyping analysis revealed multiple structural abnormalities, including chromatid exchanges and premature centromere divisions. Representative cell karyotype in G: 40,XX,chte(2;5)(F1;C2),chte(9;12)(F1;E),pcd(16)(A)[1]. (I) Representative fluorescence-activated cell sorter plots from spleen cells from a Brca1+/+ (B7292) and a Brca1−/− (B7386) mouse stained with antibodies against CD71 and Ter119. (J) Average proportion of spleen cells accumulating in regions I, II, III, and IV of red blood cell differentiation after staining with antibodies against CD71 and Ter119. Student t test was used to analyze the differences in the proportion of cells within each region. (K) Sensitivity of Brca1−/− cells (blue) relative to Brca1+/+ cells (red) in methylcellulose colony-forming assays to mitomycin C at doses of 0, 5, 10, 25, and 50 nM. Averages are shown with standard error of the mean. chrb, chromosome break; chrg, chromosome gap; chtb, chromatid break; chte, chromatid exchange; chtg, chromatid gap; pcd, premature centromere division.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/3/10.1182_blood-2015-03-635599/4/m_310f1.jpeg?Expires=1769570946&Signature=aJFO6H~6KkGTbh~-sLZUaNDRGw4Scnh2j~rM7jwtAqtqFOTkVwj-O46GPiDmT~rEDReoCWUS3~r0dOj6jU9wBvIhsR5mI1QRfQDymTHj4jymFGIsSqAMRKwmt-iVi~ZBvqzP~0NzIM-qtDsIvVQ31sm-0MzAeP0sMrbbwPLP1WHvPpvlyz4TlC1ZHLKDXt7qTv2731RyUGXJi0EpPGGcc6mUje3HogkE4Y-pOks83rjFOufMsrlsGfHaW~IrL3DW2JarYuuB0GFE~Wjq3IMXSk9NNZzn6G3kjA~s8-MjEetxqTyXtpjU3PHFcA40n~VcFPJssWGQQ1bvUTiV~UehOw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)