Key Points
eIF4E, a protein highly elevated in poor-prognostic lymphomas, simultaneously sustains expression of known driver oncogenes BCL6, BCL2, MYC.
The tumorigenic form of Hsp90 is a novel partner protein in the process underlying a new therapeutic strategy for these aggressive lymphomas.
Abstract
Aggressive double- and triple-hit (DH/TH) diffuse large B-cell lymphomas (DLBCLs) feature activation of Hsp90 stress pathways. Herein, we show that Hsp90 controls posttranscriptional dynamics of key messenger RNA (mRNA) species including those encoding BCL6, MYC, and BCL2. Using a proteomics approach, we found that Hsp90 binds to and maintains activity of eIF4E. eIF4E drives nuclear export and translation of BCL6, MYC, and BCL2 mRNA. eIF4E RNA-immunoprecipitation sequencing in DLBCL suggests that nuclear eIF4E controls an extended program that includes B-cell receptor signaling, cellular metabolism, and epigenetic regulation. Accordingly, eIF4E was required for survival of DLBCL including the most aggressive subtypes, DH/TH lymphomas. Indeed, eIF4E inhibition induces tumor regression in cell line and patient-derived tumorgrafts of TH-DLBCL, even in the presence of elevated Hsp90 activity. Targeting Hsp90 is typically limited by counterregulatory elevation of Hsp70B, which induces resistance to Hsp90 inhibitors. Surprisingly, we identify Hsp70 mRNA as an eIF4E target. In this way, eIF4E inhibition can overcome drug resistance to Hsp90 inhibitors. Accordingly, rational combinatorial inhibition of eIF4E and Hsp90 inhibitors resulted in cooperative antilymphoma activity in DH/TH DLBCL in vitro and in vivo.
Introduction
Approximately one-third of patients with diffuse large B-cell lymphoma (DLBCL) have disease that is either refractory or relapses after combinatorial chemo-immunotherapy.1,2 Mutation and constitutive expression of sets of key oncoproteins define DLBCL patients with particularly poor outcome. Among these patients, those with high expression or amplification of MYC (V-Myc avian myelocytomatosis viral oncogene homolog) show the worst outcome with an overall survival below 30% at 2 years.3-5 Frequently, MYC abnormalities are associated with either BCL2 (B-cell CLL/lymphoma 2) and/or BCL6 (B-cell CLL/lymphoma 6) mutations leading to elevated levels of these proteins.6 Almost 60% of patients with BCL2 and MYC translocations die within 6 months of diagnosis because of chemorefractory disease, a prognosis that cannot be overcome with intensified chemotherapy.5 A further hindrance to the development of new treatment regimens is the fact that these double- and triple-hit (DH/TH) lymphomas are frequently found in the elderly7 who have limited tolerability to chemotherapeutic regimens. However, novel targeted therapies disrupting key DH/TH DLBCL driver mechanisms offer for the first time opportunities to change the devastating natural history of this disease.
Previous reports indicated that the fraction of a stress active form of Hsp90 that is enriched in tumor cells (herein, tumor-enriched Hsp90 [TEHsp90]) plays an important role in lymphomagenesis.8 TEHsp90 interacts with many proteins and mediates a diverse set of mechanisms beyond its chaperone function.9,10 For example, TEHsp90 maintains the stability of BCL6 messenger RNA (mRNA) and protein, thus enabling sustained expression of BCL6 in DLBCL.8 A recently developed small molecule called PU-H71 preferentially inhibits TEHsp90 with relatively less activity against the housekeeping pool of bulk Hsp90 protein.8,11,12 Hence, PU-H71 is selectively toxic to tumor cells that are TEHsp90 dependent while sparing normal tissue.8,11,12
TEHsp90 tends to selectively bind to those proteins that are most critical for maintaining the survival of tumor cells. The small molecule PU-H71 binds tightly to TEHsp90 and locks it into its partner protein-bound configuration.13 Hence the PU-H71 molecule can serve as the basis for an affinity-capture proteomics strategy to identify TEHsp90 partner proteins that play crucial roles in cancer biology.13,14 Using this strategy, we recently mapped the TEHsp90 interactome in DLBCLs and found that several proteins regulating RNA metabolism, including eIF4E (eukaryotic translation initiation factor 4E), are part of this TEHsp90-orchestrated network of proteins required to sustain the lymphoma phenotype.12 eIF4E is a key oncogenic factor in B-cell lymphomagenesis.15 The oncogenic potential of eIF4E arises from its critical roles in the cytoplasm in the mRNA translation and in the nucleus in the mRNA export of a specific subset of transcripts.15-18 These transcripts can be regulated at the cytoplasmic (ie, translation), nuclear (ie, export), or at both levels.18 Nuclear targets are exported in the presence of eIF4E, LRPPRC (leucine-rich pentatricopeptide repeat containing), and XPO1 (exportin 1).10 eIF4E competitive inhibitors, such as ribavirin, abrogate its prosurvival function and cause antitumoral effect in solid tumors and acute myeloid leukemia (AML).19,20
Here, we show that TEHsp90 controls posttranscriptional dynamics of key mRNA species including those encoding BCL6, MYC, and BCL2 in DH/TH DLBCLs. We identify that eIF4E simultaneously modulates the mRNA export and translation of these genes and that TEHsp90 modulates eIF4E activity. We observe that eIF4E inhibition potently suppresses tumor growth through its effects on these transcripts. We also identify Hsp70 mRNA as an eIF4E target, and, in this way, eIF4E inhibition can overcome resistance to Hsp90 inhibitors. Accordingly, rational combination of eIF4E and TEHsp90 inhibitors resulted in cooperative antilymphoma activity in DH/TH DLBCL in vitro and in vivo, offering a potential new strategy for treating poor-outcome lymphomas.
Methods
Cell lines and reagents
DLBCL cell lines OCI-Ly1 and OCI-Ly18 were grown in 90% Iscove’s and 10% fetal calf serum medium (supplemented with penicillin G/streptomycin), and DLBCL cell lines SU-DHL6, DoHH2, Toledo, and Karpas422 were grown in 90% RPMI and 10% fetal calf serum medium (supplemented with penicillin G/streptomycin, N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid, and l-glutamine). The cell line U2OS was grown in Dulbecco’s modified Eagle medium with 10% fetal bovine serum and 1% penicillin/streptomycin. Cell lines were obtained from American Type Culture Collection, German Collection of Microorganism and Cell Cultures DSMZ, or the Ontario Cancer Institute. We conducted monthly testing for Mycoplasma sp. and other contaminants and quarterly cell identification by single-nucleotide polymorphism. Ribavirin was obtained from Kemprotec. PU-H71, PU-H71-beads, and control beads were synthesized and prepared at Memorial Sloan Kettering Cancer Center (T.T. and G.C.). For analysis of mRNA export, protein levels and polysomal profiling cells were treated with PU-H71 0.5 μM for 12 hours (all DLBCL cell lines), ribavirin 10 μM (OCI-Ly1 and DoHH2), or ribavirin 30 μM (SU-DHL6) for 48 hours (or 96 hours for polysomal profiling). eIF4E small-interfering RNA (siRNA) and control sequences are from Culjkovic-Kraljacic et al.16 U2OS cells were transfected with Lipofectamine (ThermoFisher) and analyzed at 72 hours. DoHH2 cells were transfected by 2 rounds of electroporation 72 hours apart (Amaxa, Lonza AG) and analyzed at 48 hours.
RIP
RNA-immunoprecipitation (RIP) from nuclear fractions of cells was performed as previously described.16 Briefly, 1 mg of nuclear lysate was used for RIP with 6 μg anti-eIF4E antibody (MBL RN001P) or control immunoglobulin G (rabbit, Millipore). After incubation, complexes were eluted by boiling in tris(hydroxymethyl)aminomethane (Tris) EDTA containing 1% sodium dodecyl sulfate and 12% β-mercaptoethanol. RNA was isolated using Trizol reagent and precipitated in isopropanol with addition of linear acrylamide. DNase-treated RNA samples (TurboDNase, Ambion) were reversed transcribed using SuperScript VILO cDNA synthesis kit (Invitrogen).
Polysomal profiling
Polysomal profiling was done as described.21 Briefly, cells were treated with cycloheximide (100 mg/mL) 10 minutes before harvesting, and lysates were prepared using polysomelysis buffer (15 mM Tris pH 7.4, 250 mM NaCl, 15 mM MgCl2, 1% Triton X-100, 100 μg/mL cycloheximide, 1 mM dithiothreitol, 400 U/mL RNaseOUT [Invitrogen], and protease inhibitors [Roche]). Equal amounts (4 mg) of protein lysates were layered on a 20% to 50% linear sucrose gradient (20% and 50% sucrose solutions in 15 mM Tris pH 7.4, 15 mM MgCl2, 150 mM NaCl, 1 mM dithiothreitol, 100 mg/μl cycloheximide, and 20 U/mL RNAseOut), mixed on Gradient Station IP Biocomp and centrifuged in a Beckman SW41Ti rotor at 92 000 G for 3 hours at 4°C. Following centrifugation, polysomal fractions were collected by continuously monitoring and recording the A254 on a Gradient Station IP (Biocomp) attached to a UV-MII (GE Healthcare) spectrophotometer. RNAs were isolated from polysomal fractions using Trizol reagent (Invitrogen).
Animal experiments
All animal procedures followed National Institutes of Health protocols and were approved by the Animal Institute Committee of the Weill Cornell Medical College.
Cell line xenografts.
SCID mice were subcutaneously injected on the right flank with DLBCL cell lines OCI-Ly1 and SU-DHL6. Tumor volumes were monitored every day using electronic calipers (Fischer Scientific). When tumor reached a palpable size (∼75 to 100 mm3), animals were randomized into 4 groups of 10 mice and treated intraperitoneally with vehicle, ribavirin, PU-H71, or the combination of ribavirin and PU-H71. We used the area-under-the-curve (AUC) as the quantitative metric to represent the evolution of tumor volume over the time frame (10 days) of the experiment. The comparisons between treated and control mice were done using multivariate analysis of variance followed by pairwise comparison using the 2-tailed Student t test (Statistix; Analytical Software, Tallahassee, FL).
Patient-derived tumorgrafts.
NSG mice were subcutaneously injected in the flank with primary human DLBCL cells. When tumor reached palpable size (as described previously), animals were randomized into 2 groups of mice and received vehicle or ribavirin 80 mg/kg intraperitoneally for 10 days. The measurement of tumor volume and comparisons between treated and control mice were performed as described previously.
Growth inhibition determination, immunohistochemistry and fluorescence in situ hybridization (FISH), RNA nuclear export, immunoblotting, metabolic labeling and capture of newly synthesized protein, RNA-sequencing (RNA-seq) and analysis, and TEHsp90 proteomics and validations are described in the supplemental Data (available on the Blood Web site). The Gene Expression Omnibus accession number for the mRNA-seq data sets reported is GSE63265.
Results
TEHsp90 proteomics points to eIF4E as a key driver of DH/TH DLBCL
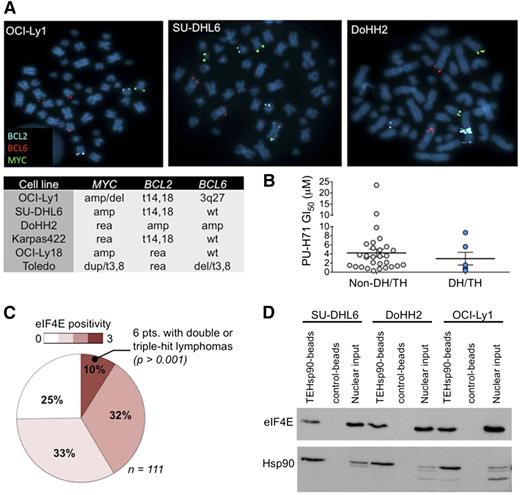
We first identified DH/TH DLBCL cell lines by FISH for MYC, BCL6, and BCL2. In a panel of 35 DLBCL cell lines, we found 6 with DH or TH mutations (OCI-Ly1, SU-DHL6, DoHH2, Karpas422, OCI-Ly18, and Toledo) (Figure 1A). Hsp90 is universally overexpressed in DLBCL specimens and to a similar extent in DH/TH and non-DH/TH cells.8 Accordingly, the activity of PU-H71 in DH/TH DLBCL is similar to non-DH/TH DLBCL cell lines (Figure 1B). This suggests that TEHsp90 may carry out diverse functions required to maintain survival also in these highly aggressive lymphoma cells. To elucidate these functions, we used PU-H71 as a chemical capture reagent to isolate TEHsp90 complexes from cell lysates of the TH lymphoma cell line OCI-Ly1 (MYC amplification, t14,18, and 3q27) by affinity purification followed by mass spectrometry. Proteomic analysis revealed that TEHsp90 was strongly bound to eIF4E (supplemental Figure 1), a critical factor in B-cell lymphomagenesis.15 Other ribosomal proteins and components of the translational machinery such as eIF4A1 were also enriched (supplemental Figure 1).
eIF4E is expressed in DH/TH DLBCL. (A) Representative image of BCL2, BCL6, and MYC FISH assay carried out in OCI-Ly1, SU-DHL6, and DoHH2 cell lines. Summary of FISH findings for MYC, BCL2, and/or BCL6 in 6 DLBCL cell lines harboring >1 of such chromosomal abnormalities. (B) Scatter plot for PU-H71 GI50 (growth inhibitory concentration 50%) in DH/TH cell lines (n = 6) vs non-DH/TH DLBCL cell lines (n = 29). (C) eIF4E expression by immunohistochemistry from 111 DLBCL cases. Number of positive cells (over total cells) is represented by color scale. All the DH/TH cases in the cohort (n = 6) presented the highest levels of eIF4E staining (n = 10). (D) Affinity purification of eIF4E and TEHsp90 from DH/TH cell lines nuclear lysates using PU-H71-beads vs chemical control beads.
eIF4E is expressed in DH/TH DLBCL. (A) Representative image of BCL2, BCL6, and MYC FISH assay carried out in OCI-Ly1, SU-DHL6, and DoHH2 cell lines. Summary of FISH findings for MYC, BCL2, and/or BCL6 in 6 DLBCL cell lines harboring >1 of such chromosomal abnormalities. (B) Scatter plot for PU-H71 GI50 (growth inhibitory concentration 50%) in DH/TH cell lines (n = 6) vs non-DH/TH DLBCL cell lines (n = 29). (C) eIF4E expression by immunohistochemistry from 111 DLBCL cases. Number of positive cells (over total cells) is represented by color scale. All the DH/TH cases in the cohort (n = 6) presented the highest levels of eIF4E staining (n = 10). (D) Affinity purification of eIF4E and TEHsp90 from DH/TH cell lines nuclear lysates using PU-H71-beads vs chemical control beads.
The dual nuclear and cytoplasmic localization of eIF4E suggested it might be active in both cellular compartments.16,18 This is of interest because eIF4E is known to mediate nuclear export of mRNAs in addition to its role in facilitating translation in the cytoplasm.16,22 Therefore, we next assessed eIF4E expression and cellular localization patterns in primary human DH/TH DLBCL patients. We analyzed eIF4E expression in 111 cases of DLBCLs by immunohistochemistry and found that in 75% of cases (n = 83) the protein was detectable under the conditions used (positive cases) (Figure 1C and supplemental Figure 2). In 72% of positive cases (n = 60 specimens), eIF4E was localized in both nuclei and cytoplasm, whereas in 10% (n = 8) and 18% (n = 15) eIF4E was exclusively detected in either the cytosolic or nuclear compartment, respectively. We then ranked nuclear and cytosolic positive cases according to the number of lymphoma cells expressing eIF4E into high expression (>60% positive lymphoma cells), midexpression (<60 and >30% positive lymphoma cells), and low expression (<30% positive lymphoma cells) (Figure 1C and supplemental Figure 2). We found that from the cohort of patients expressing high levels of eIF4E (n = 11), 6 patients also had DH/TH lymphomas defined as translocations or amplifications in MYC and BCL2 and/or BCL6 by FISH analysis (Figure 1C). There were no DH/TH lymphomas (by FISH) in patients expressing a low level or midlevel of eIF4E (P > .001 vs high eIF4E-expressing cases, Fisher’s exact test; Figure 1C). These findings suggest potential functional relevance of eIF4E in DH/TH DLBCL. Because the role of nuclear eIF4E has not yet been characterized in DH/TH DLBCL cells,18 we analyzed the association of TEHsp90 and eIF4E in this compartment. We therefore performed affinity purification of the nuclear TEHSP90 complex using PU-H71 beads and found the presence of eIF4E in the nuclear lysates of OCI-Ly1 (56% of total input), DoHH2 (66% of total input), and SU-DHL6 (54% of total input) DH/TH cell lines (Figure 1D and supplemental Figure 3), suggesting that TEHSp90 and eIF4E form a functional complex in the nuclei of DH/TH cells.
eIF4E activity regulates the nuclear export of BCL6, MYC, and BCL2
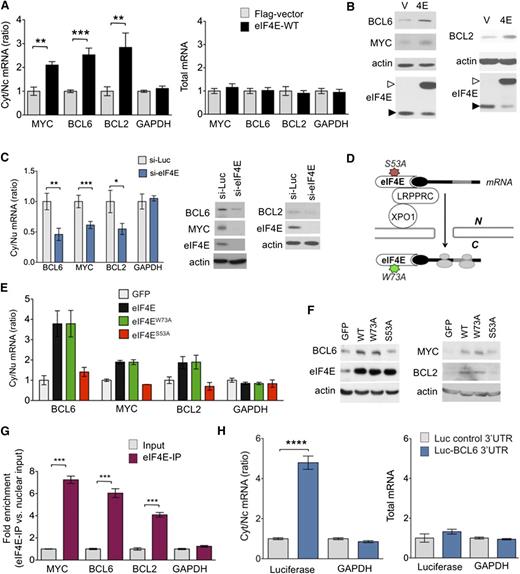
The association of high eIF4E expression and its nuclear localization in DH/TH lymphomas led us to wonder whether it could simultaneously regulate the nuclear export of BCL6, MYC, and BCL2 mRNA. MYC and BCL2 were previously described as eIF4E targets.16,23-25 To mechanistically address whether BCL6 is a nuclear export target, we took advantage of a validated cell-based assay in which U2OS cells are transfected with an eIF4E-expressing plasmid.23 The overexpression of eIF4E significantly increased the cytoplasmic to nuclear ratios of MYC and BCL2, as expected (P = .001 and P = .007, respectively; Figure 2A), and also of BCL6 (P = .0008; Figure 2A), with no effects on GAPDH. This translates into increased protein expression of MYC, BCL2, and BCL6 (Figure 2B). The effect of eIF4E on MYC, BCL2, and BCL6 nuclear RNA export was validated in reciprocal experiments using siRNA-mediated knockdown of eIF4E in U2OS cells (Figure 2C). This procedure resulted in a significantly reduced cytoplasmic to nuclear ratio of BCL6, MYC, and BCL2 mRNAs (P < .0001, P = .004, and P = .02, respectively), without changes in total cell abundance of these transcripts (Figure 2C and supplemental Figure 4A). The negative control transcript (GAPDH) was unaffected (Figure 2C). Accordingly, siRNA-mediated knockdown of eIF4E in U2OS resulted in lower levels of BCL6, MYC, and BCL2 proteins (Figure 2C). Overall, these results suggest that BCL6, BCL2, and MYC are nuclear export targets of eIF4E.
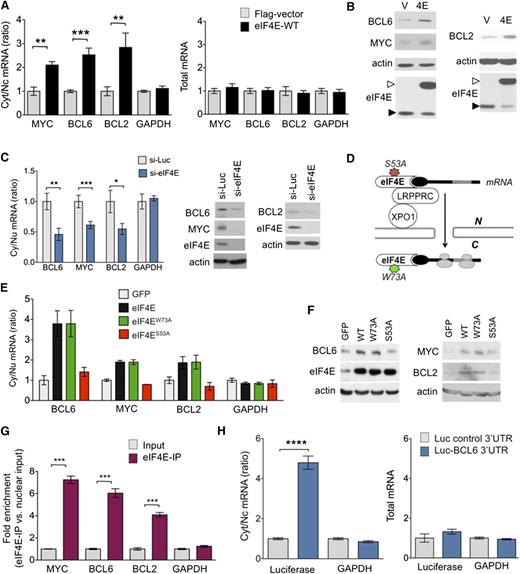
MYC, BCL2, and BCL6 are eIF4E targets. (A) Cytosolic/nuclear ratio (left) and total mRNA (right) of BCL6, BCL2, MYC, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; as control) transcripts in U2OS cells transfected with eIF4E plasmid (eIF4E-WT) or flag-vector as control. (B) Protein levels of BCL6, BCL2, MYC, actin (as control), and eIF4E in U2OS cells transfected with eIF4E plasmid (4E) or flag-vector (V). Endogenous eIF4E is marked with closed arrowhead and flagged eIF4E is marked with an open arrowhead. (C) Cytosolic/nuclear ratio of BCL6, BCL2, MYC, and GAPDH (as control) transcripts in U2OS cells transfected with siRNA for luciferase (as control) or siRNA for eIF4E. Protein levels of BCL6, BCL2, MYC, and eIF4E from the experiments are shown on the right. (D) Cartoon depicting loss of function caused by each mutant eIF4E construct used in subsequent experiment. (E) Cytosolic/nuclear ratio of BCL6, MYC, and BCL2 transcripts (GAPDH transcripts are shown as a control) and total transcript expression (right) of UOS2 cells transfected with green fluorescent protein (GFP) (as a vector control), wild-type eIF4E, and mutant eIF4E constructs eIF4EW73A and eIF4ES53A. (F) BCL6, MYC, BCL2, and eIF4E protein expression for cells shown in panel E. (G) eIF4E RIP fold enrichment of MYC, BCL2, BCL6, and GAPDH (negative control) mRNAs over input in U2OS cells. (H) Cytosolic/nuclear ratio of luciferase and GAPDH (as negative control) in U2OS cells transfected with luciferase gene with or without BCL6 3′UTR sequence. Total mRNA for the same conditions is shown on the right. ****P < .0001.
MYC, BCL2, and BCL6 are eIF4E targets. (A) Cytosolic/nuclear ratio (left) and total mRNA (right) of BCL6, BCL2, MYC, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; as control) transcripts in U2OS cells transfected with eIF4E plasmid (eIF4E-WT) or flag-vector as control. (B) Protein levels of BCL6, BCL2, MYC, actin (as control), and eIF4E in U2OS cells transfected with eIF4E plasmid (4E) or flag-vector (V). Endogenous eIF4E is marked with closed arrowhead and flagged eIF4E is marked with an open arrowhead. (C) Cytosolic/nuclear ratio of BCL6, BCL2, MYC, and GAPDH (as control) transcripts in U2OS cells transfected with siRNA for luciferase (as control) or siRNA for eIF4E. Protein levels of BCL6, BCL2, MYC, and eIF4E from the experiments are shown on the right. (D) Cartoon depicting loss of function caused by each mutant eIF4E construct used in subsequent experiment. (E) Cytosolic/nuclear ratio of BCL6, MYC, and BCL2 transcripts (GAPDH transcripts are shown as a control) and total transcript expression (right) of UOS2 cells transfected with green fluorescent protein (GFP) (as a vector control), wild-type eIF4E, and mutant eIF4E constructs eIF4EW73A and eIF4ES53A. (F) BCL6, MYC, BCL2, and eIF4E protein expression for cells shown in panel E. (G) eIF4E RIP fold enrichment of MYC, BCL2, BCL6, and GAPDH (negative control) mRNAs over input in U2OS cells. (H) Cytosolic/nuclear ratio of luciferase and GAPDH (as negative control) in U2OS cells transfected with luciferase gene with or without BCL6 3′UTR sequence. Total mRNA for the same conditions is shown on the right. ****P < .0001.
In order to more rigorously distinguish whether effects of eIF4E on BCL6 are due solely to nuclear export or might also be linked to more efficient translation, we performed similar experiments, but this time using an eIF4E mutant (eIF4ES53A) that disrupts its export function leaving translation intact, and an eIF4E mutant (eIF4EW73A) that disrupts its effect on translation but not mRNA transport, using U2OS cells as a tractable model for these transfection experiments (Figure 2D).16,21,23 Similar to wild-type eIF4E overexpression, the nuclear export of BCL6 mRNA was elevated to more than threefold in eIF4EW73A mutant cells relative to vector, whereas GAPDH was unaffected (Figure 2E). This corresponded to increased BCL6 protein levels (Figure 2F). On the contrary, eIF4ES53A did not increase BCL6 (or GAPDH) nuclear mRNA export or protein levels (Figure 2E-F). We observed a similar effect at the transcript and protein levels for BCL2 and MYC expression (Figure 2F). Total levels of BCL6, MYC, BCL2, and GAPDH mRNA were not changed in cells transduced with eIF4E mutants (supplemental Figure 4B).
To directly determine the binding of BCL6 mRNA to nuclear eIF4E, we carried out eIF4E immunoprecipitation from nuclear fractions followed by quantitative polymerase chain reaction (RIP-qPCR). We found that BCL6 transcripts, like BCL2 and MYC, were highly enriched in eIF4E RIPs relative to nuclear input (P < .0001 for all genes; Figure 2G), suggesting a direct binding of eIF4E to BCL6 mRNA. RNAs exported by eIF4E contain a 50-nucleotide element in the 3′ untranslated region (UTR) known as a 4E-SE element.16 We found one of these elements present in the 3′UTR of BCL6. To determine the role of the 3′UTR in the mRNA export of BCL6, we transfected U2OS cells overexpressing wild-type eIF4E with a construct expressing luciferase fused to the 3′UTR of BCL6 (luc-BCL6) or luciferase only control and monitored the export of luciferase transcripts. Consistently, eIF4E overexpression leads to a significantly increased export of luc-BCL6 but not luciferase alone (P < .0001; Figure 2H), whereas total mRNA levels of luc-BCL6 were not affected (Figure 2H). Importantly, there was no effect on endogenous GAPDH mRNAs (negative control), which are not eIF4E targets (Figure 2H). BCL6 is thus a direct mRNA export target of eIF4E. Our data suggest eIF4E regulates BCL6 expression through increased mRNA export but not through more efficient translation. To further investigate this, we carried out polysomal fractionation in U2OS cells overexpressing eIF4E or control GFP and analyzed BCL6 mRNA in each fraction by qPCR. Here, BCL6 exhibited only a modest shift to higher-molecular-weight polysomes in the presence of eIF4E. In contrast, vascular endothelial growth factor A, an established translational target of eIF4E,26 manifested a substantial polysomal shift (supplemental Figure 5A-C). These results further underline that eIF4E facilitates BCL6 protein expression mainly through nuclear export of its mRNA, at least in this cell type.
Nuclear eIF4E controls an extended oncogenic program in lymphoma cells
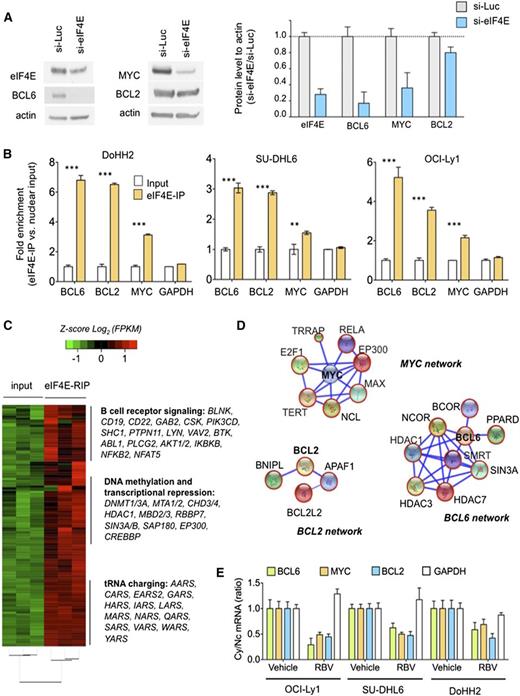
The nuclear targets of eIF4E are cell-type specific. We therefore determined whether eIF4E regulates BCL6, MYC, and BCL2 transcripts in the context of DH/TH DLBCL cells. Similar to U2OS cells, siRNA-mediated knockdown of eIF4E in the TH DoHH2 cell line resulted in lower levels of BCL6, MYC, and BCL2 proteins (Figure 3A and supplemental Figure 6). We then performed RIP-qPCR in nuclear extracts of DoHH2, OCI-Ly1, and SU-DHL6 DH/TH cell lines. We found that BCL6, BCL2, and MYC transcripts were significantly enriched compared with control mRNA in eIF4E RIPs relative to input in OCI-Ly1 (P < .0001 for all genes; Figure 3B), SU-DHL6 cells (P < .0001, P = .005, and P < .0001, respectively; Figure 3B), and DoHH2 cells (P < .0001 for all genes; Figure 3B) indicating that nuclear eIF4E binds to these transcripts in this specific cellular context. There were no significant changes in GAPDH transcripts used as negative control (Figure 3B).
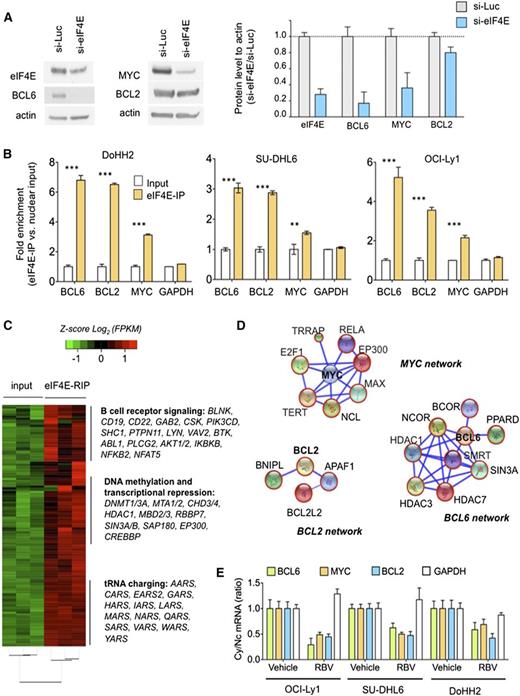
Nuclear eIF4E regulates the export of lymphomagenic transcripts. (A) Protein levels of BCL6, BCL2, MYC, and eIF4E in the DH/TH DoHH2 cells transfected with siRNA for luciferase (as control) or siRNA for eIF4E. Protein quantification is shown on the right for triplicate experiments as mean ± standard error of the mean (SEM). (B) eI4FE mRNA immunoprecipitation for BCL6, MYC, BCL2, and GAPDH (as control) in DoHH2, SU-DHL6, and OCI-Ly1 nuclear fractions. Results expressed as fold enrichment over nuclear input. ***P < .001 and **P < .05 (Student t test for triplicates). (C) eI4FE RIP sequencing (RIP-seq) in OCI-Ly1 cells (vs nuclear input). Selected transcripts and pathways differentially enriched in eIF4E RIPs are shown on the right. (D) eIF4E nuclear targets that sustain BCL6, MYC, and BCL2 activity in lymphoma cells depicted as “networks” from Search Tool for the Retrieval of Interacting Genes/Proteins analysis. Transcripts identified in this eIF4E RIP-seq are shown in red circles. (E) Effect of ribavirin (RBV) on the mRNA nuclear export (determined by mRNA cytosolic/nuclear ratio) of BCL6, MYC, BCL2, and GAPDH (as control) in SU-DHL6, OCI-Ly1, and DoHH2 cells.
Nuclear eIF4E regulates the export of lymphomagenic transcripts. (A) Protein levels of BCL6, BCL2, MYC, and eIF4E in the DH/TH DoHH2 cells transfected with siRNA for luciferase (as control) or siRNA for eIF4E. Protein quantification is shown on the right for triplicate experiments as mean ± standard error of the mean (SEM). (B) eI4FE mRNA immunoprecipitation for BCL6, MYC, BCL2, and GAPDH (as control) in DoHH2, SU-DHL6, and OCI-Ly1 nuclear fractions. Results expressed as fold enrichment over nuclear input. ***P < .001 and **P < .05 (Student t test for triplicates). (C) eI4FE RIP sequencing (RIP-seq) in OCI-Ly1 cells (vs nuclear input). Selected transcripts and pathways differentially enriched in eIF4E RIPs are shown on the right. (D) eIF4E nuclear targets that sustain BCL6, MYC, and BCL2 activity in lymphoma cells depicted as “networks” from Search Tool for the Retrieval of Interacting Genes/Proteins analysis. Transcripts identified in this eIF4E RIP-seq are shown in red circles. (E) Effect of ribavirin (RBV) on the mRNA nuclear export (determined by mRNA cytosolic/nuclear ratio) of BCL6, MYC, BCL2, and GAPDH (as control) in SU-DHL6, OCI-Ly1, and DoHH2 cells.
To determine the extent of nuclear eIF4E activity in DH/TH DLBCLs and how these programs can support the oncogenic activity of BCL6, MYC, and/or BCL2 transcripts, we conducted eIF4E RIP of nuclear RNA followed by RNA-seq in OCI-Ly1 cells in biological triplicates. Compared with the nuclear input, eIF4E binding was significantly enriched in 3958 transcripts (Figure 3C and supplemental Table 1). BCL6, MYC, and BCL2 were enriched 5.4-, 1.2-, and 2.7-fold, respectively, by this method. Pathways analysis suggested a role of eIF4E preferential nuclear cargos in processes like B-cell receptor signaling (eg, AKT, BTK, BCL6, CD19, CD22, CSK, NFAT, RELA, SHC1), DNA methylation and epigenetic regulation (eg, DNMT1, DNMT3A, HDAC1, MBD2, MBD3), and transfer RNA charging/metabolism (eg, AARS, CARS, HARS, LARS) (Figure 3C and supplemental Figure 7), overall indicating that eIF4E regulates a vast network of lymphoma-sustaining pathways. eIF4E target transcripts include components of BCL6 corepressor complexes like SMRT, NCOR, BCOR, HDAC1, HDAC3, HDAC7, PPARD, and SIN3A (Figure 3D); MYC transcriptional complexes like MAX, EP300, RELA, YY1AP1, E2F1, NCL, TERT, and TRRAP (Figure 3D); and BCL2 interacting proteins like BCL-W (BCL2L2), APAF1, and BNIPL (Figure 3D).
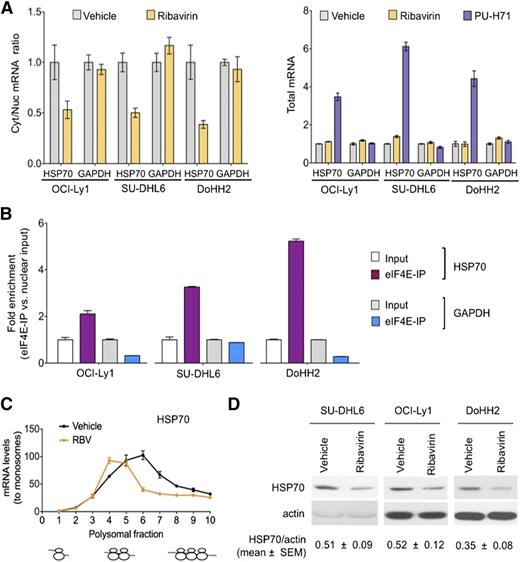
We next examined whether disruption of eIF4E binding to BCL6, MYC, and BCL2 transcripts could impair their export and preferential translation in the DH/TH cell lines OCI-Ly1, SU-DHL6, and DoHH2. For these studies, we used the small molecule ribavirin, which competes for the methyl-cap binding site and so prevents eIF4E from binding to its mRNA targets.22 We found that ribavirin significantly decreased cytosolic to nuclear ratios of BCL6, MYC, and BCL2 mRNA, but not of GAPDH, in all the cell lines tested (Figure 3E).
TH lymphomagenic transcripts are simultaneously targeted by ribavirin in vitro and in vivo
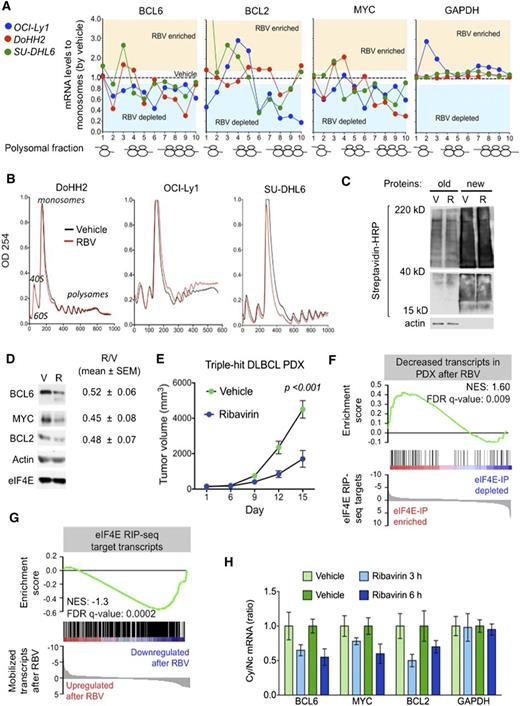
The data discussed above indicate that ribavirin decreases the nuclear export of BCL6, MYC, and BCL2 in DH/TH lymphoma cells overexpressing these transcripts. To determine the effect of eIF4E inhibition on the cytosolic translation of these genes, we exposed OCI-Ly1, DoHH2, and SU-DHL6 cells to ribavirin followed by polysomal fractionation and qPCR. We found that ribavirin depleted these mRNAs (but not GAPDH) from the higher-molecular-weight polysomal fractions (Figure 4A and supplemental Figure 8A). Importantly, ribavirin did not affect ribosomal assembly (Figure 4B) or change total mRNA levels in these DLBCL cell lines (supplemental Figure 8B). Ribavirin did not impact global protein production as shown in the TH cell line OCI-Ly1 (Figure 4C). Overall, these data suggest that in the context of DH/TH lymphoma cells, eIF4E influences both nuclear export and translation of oncogenic transcripts. Accordingly, ribavirin treatment of 48 hours resulted in reduced protein levels of BCL6, MYC, and BCL2 in the DH/TH DLBCL cell lines SU-DHL6 (Figure 4D, blot), OCI-Ly1, and DoHH2 (Figure 4D, blots shown in Figure 5A and supplemental Figure 10). Is noteworthy that, unlike U2OS cells, we also observed translational control of these transcripts in lymphoma cells highlighting the relevance of context to this type of regulation.
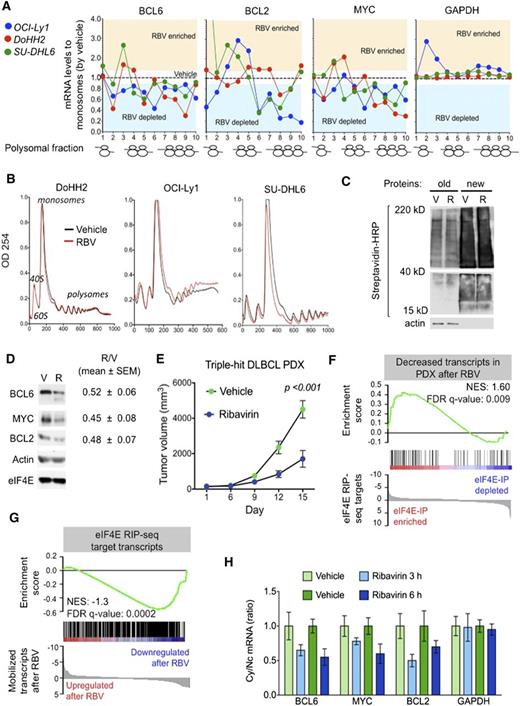
Ribavirin is active a TH DLBCL patient-derived xenograft (PDX) model. (A) Polysomal profiling of BCL6, BCL2, MYC, and GAPDH (as control) transcripts in OCI-Ly1, DoHH2, and SU-DHL6 cells treated with vehicle (black dotted line) vs ribavirin (RBV). Each profiling is normalized to the respective vehicle represented with a dotted central line. Points localized above and below the line represent polysome fractions enriched and depleted in RBV-treated cells. (B) Total polysomal profiling of DoHH2, OCI-Ly1, and SU-DHL6 cells treated with vehicle (black line) vs ribavirin (red line). (C) Total protein levels in OCI-Ly1 cells treated with vehicle (V) and ribavirin (R). Previously synthesized protein is marked as old and newly synthesized protein is marked as new. Actin was used as control. (D) Protein levels of BCL6, BCL2, MYC, eIF4E, and actin (as control) in SU-DHL6 cells treated with vehicle (V) vs ribavirin (R) for 48 hours. Densitometry analysis is shown on the right for replicates experiments as mean ± SEM conducted in SU-DHL6, OCI-Ly1, and DoHH2 cell lines. (E) In vivo effect of ribavirin (blue dots) vs vehicle (green dots) in the PDX-4 mice. (F-G) Gene Set Enrichment Analysis of decreased transcripts upon ribavirin treatment in PDX-4 mice and eIF4E nuclear targets from RIP-seq experiments. (H) Cytosolic vs nuclear distribution of BCL6, BCL2, MYC, and GAPDH (as control) mRNAs from the PDX-4 after ribavirin treatment of 3 hours or 6 hours. FDR, false discovery rate; NES, normalized enrichment score.
Ribavirin is active a TH DLBCL patient-derived xenograft (PDX) model. (A) Polysomal profiling of BCL6, BCL2, MYC, and GAPDH (as control) transcripts in OCI-Ly1, DoHH2, and SU-DHL6 cells treated with vehicle (black dotted line) vs ribavirin (RBV). Each profiling is normalized to the respective vehicle represented with a dotted central line. Points localized above and below the line represent polysome fractions enriched and depleted in RBV-treated cells. (B) Total polysomal profiling of DoHH2, OCI-Ly1, and SU-DHL6 cells treated with vehicle (black line) vs ribavirin (red line). (C) Total protein levels in OCI-Ly1 cells treated with vehicle (V) and ribavirin (R). Previously synthesized protein is marked as old and newly synthesized protein is marked as new. Actin was used as control. (D) Protein levels of BCL6, BCL2, MYC, eIF4E, and actin (as control) in SU-DHL6 cells treated with vehicle (V) vs ribavirin (R) for 48 hours. Densitometry analysis is shown on the right for replicates experiments as mean ± SEM conducted in SU-DHL6, OCI-Ly1, and DoHH2 cell lines. (E) In vivo effect of ribavirin (blue dots) vs vehicle (green dots) in the PDX-4 mice. (F-G) Gene Set Enrichment Analysis of decreased transcripts upon ribavirin treatment in PDX-4 mice and eIF4E nuclear targets from RIP-seq experiments. (H) Cytosolic vs nuclear distribution of BCL6, BCL2, MYC, and GAPDH (as control) mRNAs from the PDX-4 after ribavirin treatment of 3 hours or 6 hours. FDR, false discovery rate; NES, normalized enrichment score.
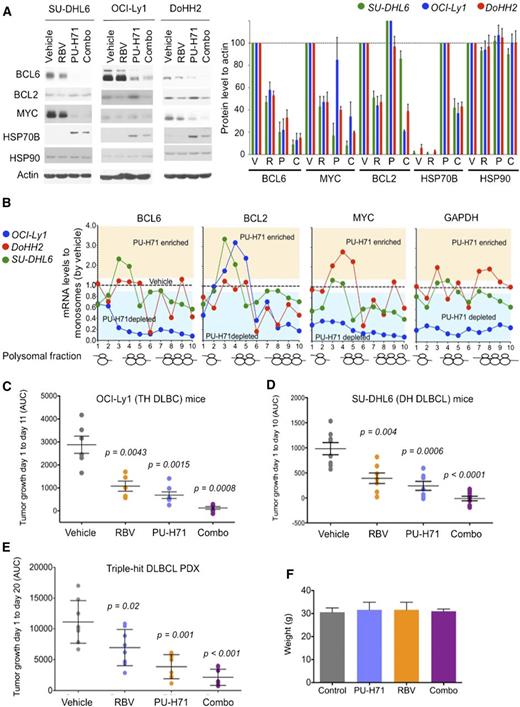
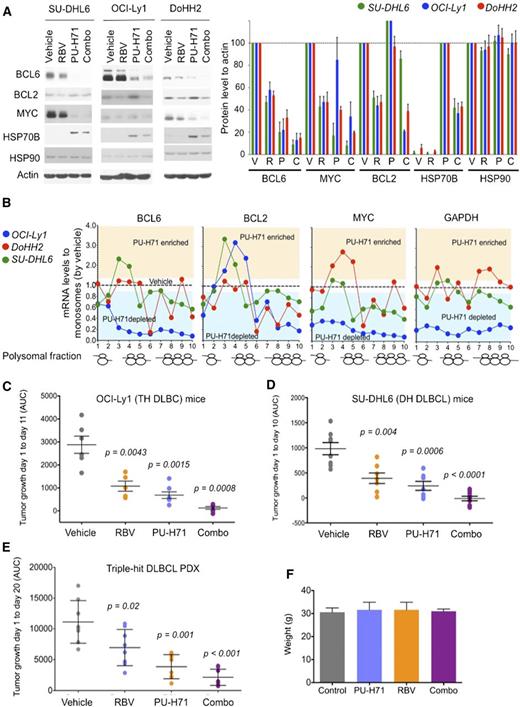
Antilymphoma effect of combined inhibition of eIF4E and TEHsp90. (A) Effect on BCL6, BCL2, MYC, Hsp70, Hsp90, and actin (as control) protein abundance of vehicle, ribavirin (RBV), PU-H71, and their combination (combo) in SU-DHL6 (SU6), DoHH2 (Do2), and OCI-Ly1 (Ly1) cells. Densitometry analysis is shown on the right for triplicate experiments as mean ± SEM. C, combination; P, PU-H71; R, ribavirin; V, vehicle. (B) BCL6, MYC, BCL2, and GAPDH polysomal profiling of OCI-Ly1, DoHH2, and SU-DHL6 cells treated with vehicle (black dotted line) or the TEHsp90 inhibitor PU-H71. Each profiling is normalized to its respective vehicle represented with a dotted central line. Points localized below the line represent polysome fractions depleted in PU-H71-treated cells. (C-D) Tumor growth (in AUC from day 1 to day 10) of OCI-Ly1 and SU-DHL6 xenografted mice. P values are shown on top for each group for the comparison with vehicle-treated mice. (E) Tumor growth (in AUC from day 1 to day 20) of TH DLBCL PDX xenografted mice. P values are shown on top for each group for the comparison with vehicle-treated mice. (F) Body weight at day 21 of mice from panel E.
Antilymphoma effect of combined inhibition of eIF4E and TEHsp90. (A) Effect on BCL6, BCL2, MYC, Hsp70, Hsp90, and actin (as control) protein abundance of vehicle, ribavirin (RBV), PU-H71, and their combination (combo) in SU-DHL6 (SU6), DoHH2 (Do2), and OCI-Ly1 (Ly1) cells. Densitometry analysis is shown on the right for triplicate experiments as mean ± SEM. C, combination; P, PU-H71; R, ribavirin; V, vehicle. (B) BCL6, MYC, BCL2, and GAPDH polysomal profiling of OCI-Ly1, DoHH2, and SU-DHL6 cells treated with vehicle (black dotted line) or the TEHsp90 inhibitor PU-H71. Each profiling is normalized to its respective vehicle represented with a dotted central line. Points localized below the line represent polysome fractions depleted in PU-H71-treated cells. (C-D) Tumor growth (in AUC from day 1 to day 10) of OCI-Ly1 and SU-DHL6 xenografted mice. P values are shown on top for each group for the comparison with vehicle-treated mice. (E) Tumor growth (in AUC from day 1 to day 20) of TH DLBCL PDX xenografted mice. P values are shown on top for each group for the comparison with vehicle-treated mice. (F) Body weight at day 21 of mice from panel E.
To assess the antilymphoma effect of eIF4E inhibition, we established a PDX in NSG mice. The specimen was isolated from a newly diagnosed patient harboring a TH DLBCL. The patient presented a stage IVb, International Prognostic Index 4 disease with bone marrow infiltration and was refractory to multiple therapeutic regimens (supplemental Figure 9A). Once engrafted in NSG mice, the specimen was followed by FISH and immunoblotting for MYC, BCL2, and BCL6 to ensure it remained a TH disease with similar expression levels of these proteins (supplemental Figure 9A). The fourth generation of PDX (PDX-4) was tested ex vivo for doxorubicin and ribavirin response. Once we proved PDX-4 was chemoresistant to doxorubicin and responsive to ribavirin ex vivo (supplemental Figure 9B), we expanded into NSG mice for preclinical therapeutic assessment. To determine whether primary human DLBCL is eIF4E dependent in vivo, we implanted PDX-4 tumors into the flanks of 10 NSG mice, and when tumors were palpable (75-100 mm3), mice were randomized to receive vehicle or ribavirin 80 mg/kg twice daily intraperitoneally for 10 days. We found a significant reduction in tumor growth in PDX-4 after ribavirin treatment (P < .001, Student t test; Figure 4E) with no signs of toxicity by weight or by macroscopic and microscopic examination of organs including liver, kidney, lungs, bone marrow, and heart. We then analyzed the transcriptome in vehicle vs ribavirin-treated PDX-4 (at day 10) by RNA-seq followed by Gene Set Enrichment Analysis. We found that significantly decreased transcripts in PDX-4 after ribavirin were significantly enriched in eIF4E RIP-seq targets obtained from the TH cell line OCI-Ly1 (NES, 1.60; FDR q value, 0.009; Figure 4F). Conversely, eIF4E RIP-seq transcripts were characterized by decreased enrichment of these transcripts upon ribavirin treatment (NES, −1.3; FDR q value, 0.0002; Figure 4G).
To further confirm the pharmacodynamic action of ribavirin on mRNA transport in the primary human PDX setting, we examined the abundance of BCL6, BCL2, and MYC transcripts in the cytoplasm of implanted PDX-4 cells. The patient lymphoma cells were implanted into the flanks of 8 NSG recipients followed by exposure to ribavirin 80 mg/kg intraperitoneally (n = 4) for 3 and 6 hours (n = 2 mice, respectively), a clinically achievable dose in patients. An additional 4 PDX-4-implanted mice were treated in similar fashion with vehicle. We observed a decrease in the cytosolic/nuclear ratio of BCL6, BCL2, and MYC as early as 3 hours after the administration (Figure 4H).
Ribavirin and PU-H71 show cooperative antilymphoma activity in DH/TH DLBCL
Given our initial findings showing eIF4E is a partner of TEHsp90 in the nuclei of DH/TH cells, we reasoned that combining TEHsp90- and eIF4E-targeted therapies would achieve greater target inhibition and hence antilymphoma activity. We therefore compared and contrasted the effect of ribavirin, PU-H71, or the combination on the expression of BCL6, MYC, and BCL2 by immunoblotting in the DH/TH DLBCL cell lines OCI-Ly1, SU-DHL6, and DoHH2. Compared with either drug alone, the combination more potently suppressed the expression of BCL6 and MYC in all 3 cell lines (Figure 5A and supplemental Figure 10) and BCL2 in OCI-Ly1 cells (Figure 5A and supplemental Figure 10). At this time point, PU-H71 stabilizes or increases BCL2 protein expression, a known effect of Hsp90 inhibitors before inducing protein degradation.12 Next, we explored the effects of PU-H71 on mRNA translation because other components of the translational machinery complex like eIF4A1, eIF2, eIF3, and PABP were also associated with TEHsp90 in the TH DLBCL cell line OCI-Ly1 (supplemental Figure 1).12 We found that TEHsp90 inhibition rapidly promotes ribosomal disassembly (ie, decreases polysomes) while increasing 40S, 60S, and monosome fractions in OCI-Ly1, DoHH2, and SU-DHL6 cells (supplemental Figure 11). This effect was more pronounced for actively translated mRNAs like BCL6, MYC, and BCL2 in DH/TH cell lines (Figure 5B and supplemental Figure 12) and likely contributes to the decreased MYC and BCL6 protein levels observed with PU-H71 (Figure 5A). The effects on polysomal assembly are unique to TEHsp90 inhibition because ribavirin treatment does not affect polysome integrity22 (see also Figure 4B). Hence, suppression of TEHsp90 has additional effects on translation than those conferred by its interaction with eIF4E. This is also evident in the disassembling of polysomes on GAPDH in OCI-Ly1 cells (Figure 5B and supplemental Figure 12).
To examine the antilymphoma effect of this combination, we xenografted TH OCI-Ly1 cells (n = 21) and DH SU-DHL6 cells (n = 31) in SCID mice, and TH PDX-4 (n = 32) in NSG mice. Once tumors developed to a palpable size (75-100 mm3), we treated them with vehicle control (n = 6, 8, and 8 in OCI-Ly1, SU-DHL6, and PDX-4, respectively), ribavirin 40 mg/kg per day (n = 5, 7, and 8 in OCI-Ly1, SU-DHL6, and PDX-4, respectively), PU-H71 25 mg/kg per day (n = 5, 7, and 8 in OCI-Ly1, SU-DHL6, and PDX-4, respectively), or the combination (n = 5, 9, and 8 in OCI-Ly1, SU-DHL6, and PDX-4, respectively) for 10 days. Compared with vehicle-treated mice, ribavirin and PU-H71 significantly decreased lymphoma growth in OCI-Ly1 mice (P = .0043 and P = .0015, respectively; Figure 5C), SU-DHL6 mice (P = .004 and P = .0006, respectively; Figure 5D), and PDX-4 mice (P = .001 and P = .02, respectively; Figure 5E). Compared with vehicle-treated mice, the combination of ribavirin with PU-H71 suppressed lymphoma growth more profoundly in the 3 models (P = .0008 in OCI-Ly1, P < .0001 in SU-DHL6, and P < .0001 in PDX-4; Figure 5C-E). There was no macroscopic or microscopic evidence of toxicity in any mice. There were no changes in body weight by the end of the treatment (Figure 5F) or in blood biochemistry values (supplemental Figure 2).
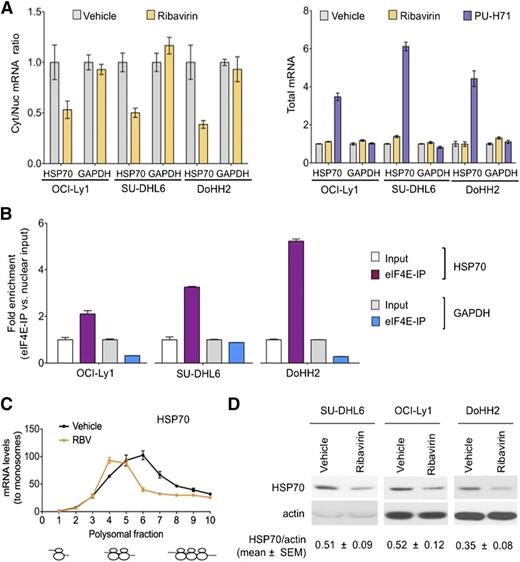
In the previous studies, we monitored Hsp70 as a control for TEHsp90 inhibition. It is well-known that Hsp70 becomes activated under these conditions.9,10 This elevation of Hsp70 induces antiapoptotic mechanisms27 leading to resistance to Hsp90 inhibitors and limiting the activity of these drugs in the clinic. Surprisingly, we noted that eIF4E inhibition reduced the increase in Hsp70 (40%-60%) (Figure 5A). Given these findings, we examined whether Hsp70 was a direct eIF4E target. Indeed, we found that ribavirin decreased the nuclear export of Hsp70 without affecting its total RNA levels in OCI-Ly1, SU-DHL6, and DoHH2 cells (Figure 6A). Accordingly, Hsp70 transcripts specifically bound eIF4E in the nuclear fraction in all these DH/TH cell lines, supporting that Hsp70 (HSPA6) mRNA is a direct eIF4E target (Figure 6B). Further, ribavirin decreased the amount of Hsp70 on heavy polysomes as demonstrated in OCI-Ly1 cells (Figure 6C). Overall, this led to a ribavirin-induced decrease in Hsp70 abundance in SU-DHL6, OCI-Ly1, and DoHH2 cells (Figure 6D). This suggests that an additional reason for the potentiation between the 2 drugs is because ribavirin treatment enhances PU-H71 activity by reducing Hsp70. Hence, the combination of ribavirin and PU-H71 enhances each other’s activity at multiple levels.
Ribavirin decreases TEHsp90 inhibition–induced Hsp70 upregulation. (A) Effect of ribavirin on the mRNA nuclear export (determined by mRNA cytosolic/nuclear ratio) of HSP70B (HSPA6) and GAPDH (as control) in OCI-Ly1, SU-DHL6, and DoHH2 cells. The effect on total mRNA is sown on the right with PU-H71 effect as control. (B) eI4FE mRNA immunoprecipitation for HSP70B and GAPDH (as control) in OCI-Ly1, SU-DHL6, and DoHH2 nuclear fractions. Results expressed as fold enrichment over nuclear input. (C) Polysomal profiling of HSP70B transcript in OCI-Ly1 cells treated with vehicle vs ribavirin (RBV). (D) Effect on Hsp70 and actin (as control) protein abundance in ribavirin- and vehicle-treated SU-DHL6, OCI-Ly1, and DoHH2 cells. Immunoblots from 50 μg of cell lysates were exposed for optimal time to visualize Hsp70 at baseline. Hsp70 protein quantification (to actin) is shown at the bottom for quadruplicate experiments as mean ± SEM.
Ribavirin decreases TEHsp90 inhibition–induced Hsp70 upregulation. (A) Effect of ribavirin on the mRNA nuclear export (determined by mRNA cytosolic/nuclear ratio) of HSP70B (HSPA6) and GAPDH (as control) in OCI-Ly1, SU-DHL6, and DoHH2 cells. The effect on total mRNA is sown on the right with PU-H71 effect as control. (B) eI4FE mRNA immunoprecipitation for HSP70B and GAPDH (as control) in OCI-Ly1, SU-DHL6, and DoHH2 nuclear fractions. Results expressed as fold enrichment over nuclear input. (C) Polysomal profiling of HSP70B transcript in OCI-Ly1 cells treated with vehicle vs ribavirin (RBV). (D) Effect on Hsp70 and actin (as control) protein abundance in ribavirin- and vehicle-treated SU-DHL6, OCI-Ly1, and DoHH2 cells. Immunoblots from 50 μg of cell lysates were exposed for optimal time to visualize Hsp70 at baseline. Hsp70 protein quantification (to actin) is shown at the bottom for quadruplicate experiments as mean ± SEM.
Discussion
Our findings provide a molecular basis for previous data showing that TEHsp90 had effects on mRNA metabolism on BCL6 and other oncogenic transcripts in DLBCL.8 We demonstrate that TEHsp90 sustains eIF4E activity in the nucleus and cytoplasm of DH/TH lymphoma cells and that, through this interaction, the oncogenic activities of Hsp90 and eIF4E are mutually reinforced. Further, we demonstrate that eIF4E elevation, through its effects on Hsp70 mRNA export and translation, likely drives Hsp90 drug resistance. Thus, eIF4E inhibition increases the potency of Hsp90 inhibitors, as we observe in our xenograft models. These findings suggest a role for eIF4E in the regulation of the cellular stress response, particularly in the oncogenic milieu. In this context, we described for the first time that nuclear eIF4E controls an extensive network of transcripts with roles in B-cell receptor signaling (BTK, CD19, CD22); folate, glucose and amino acid metabolism (MTHFD1, MTOR, PI3Ks, MAPKs, YARS, CARS); DNA repair and cellular stress (PARPs, XRCC1, RAD50, RAD52, Hsp70, Hsp40); and transcriptional regulation including chromatin remodeling and epigenetic enzymes (DNMTs, HDACs, MBDs, CHDs, EP300). These findings explain the dependence of lymphoma cells on the activity of eIF4E to fully express the transformation capabilities of classic lymphoma oncogenes such as BCL6, BCL2, and MYC that are also eIF4E targets. Therefore, by targeting eIF4E both the oncogenes and their supporting network of genes are simultaneously dismantled.
Our findings provide a strong rationale for translating eIF4E inhibitors and TEHsp90 inhibitors in the treatment of aggressive diseases such as DH/TH lymphomas. Importantly, ribavirin is well tolerated and leads to responses in patients with AML.19 In this trial, ribavirin showed single-agent activity in patients with poor-prognosis AML, including 6 patients (out of 11 evaluable for response) with remissions or blast responses.19 Micromolar plasma concentrations used here are achievable in humans with minimal toxicity.19,28,29 Similar to ribavirin, early stage trials (www.clinicaltrials.gov, #NCT01581541) indicate that the TEHsp90 inhibitor PU-H71 is safe, and thus the combination of these 2 drugs seems feasible, as our studies in mice indicate. Remarkably, by decreasing Hsp70 export and translation, ribavirin could diminish one of the most important mechanisms in the acquired resistance to this class of Hsp90 inhibitors. In summary, our data provide a novel and potentially nontoxic mechanistic-based approach to target aggressive B-cell lymphomas harboring multiple oncogene activation.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE63265).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Cristina Montagna and Jidong Shan of the Molecular Cytogenetic Core of the Albert Einstein College of Medicine for their help with FISH assays in cell lines.
This work was supported by grants from the Leukemia and Lymphoma Society (LLS 6078-14 to L.C., LLS 6478 and LLS 6160 to K.L.B.B.), the Raymond and Beverly Sackler Scholar (L.C.), and the National Institutes of Health, National Cancer Institute (grants R01 98571 and R01 80728) (K.L.B.B.). K.L.B.B. holds a Canada Research Chair.
Authorship
Contribution: B.C.-K., T.M.F., R.L.G., R. Marullo, N.C.-V., A.V., H.Z., and J.P. conducted cell-based experiments, RIP-seq, and analysis; S.Y., F.T., M.G., P.G., M.L., R. Machiorlatti, and G.I. conducted animal experiments and established PDX; T.T. and G.C. prepared and provided reagents; A.M., O.E., K.L.B.B., and L.C. supervised research and designed experiments; and B.C.-K., T.M.F., A.M., K.L.B.B., and L.C. wrote the manuscript.
Conflict-of-interest disclosure: Memorial Sloan Kettering Cancer Center holds the intellectual rights to PU-H71. Samus Therapeutics, of which G.C. has partial ownership, has licensed PU-H71. The remaining authors declare no competing financial interests.
Correspondence: Katherine L. B. Borden, Institute for Research in Immunology and Cancer and Department of Pathology and Cell Biology, Université de Montréal, Montréal, QC, Canada; e-mail: katherine.borden@umontreal.ca; and Leandro Cerchietti, Hematology and Oncology Division, Weill Cornell Medical College, 1300 York Ave, C620B, New York, NY 10065; e-mail: lec2010@med.cornell.edu.
References
Author notes
B.C.-K. and T.M.F. contributed equally to this study.