Key Points
Bortezomib induces the degradation of FLT3-ITD through an autophagy-dependent mechanism that contributes to cell death.
This finding provides a mechanism-based rationale for the study of proteasome inhibitors in FLT3-ITD–mutant acute myeloid leukemia.
Abstract
Internal tandem duplication of the Fms-like tyrosine kinase-3 receptor (FLT3) internal tandem duplication (ITD) is found in 30% of acute myeloid leukemia (AML) and is associated with a poor outcome. In addition to tyrosine kinase inhibitors, therapeutic strategies that modulate the expression of FLT3-ITD are also promising. We show that AML samples bearing FLT3-ITD mutations are more sensitive to proteasome inhibitors than wild-type samples and this sensitivity is strongly correlated with a higher FLT3-ITD allelic burden. Using pharmacologic inhibitors of autophagy, specific downregulation of key autophagy proteins including Vps34, autophagy gene (Atg)5, Atg12, Atg13, biochemical, and microscopy studies, we demonstrated that proteasome inhibitors induced cytotoxic autophagy in AML cells. FLT3-ITD molecules were detectable within autophagosomes after bortezomib treatment indicating that autophagy induction was responsible for the early degradation of FLT3-ITD, which preceded the inhibition of mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK), PI3K/AKT, and STAT5 pathways, and subsequent activation of cell death. Moreover, proteasome inhibitors overcome resistance to quizartinib induced by mutations in the kinase domain of FLT3, suggesting that these compounds may prevent the emergence of mutant clones arising from tyrosine kinase inhibitor treatments. In xenograft mice models, bortezomib stimulated the conversion of LC3-I to LC3-II, indicating induction of autophagy in vivo, downregulated FLT3-ITD protein expression and improved overall survival. Therefore, selecting patients according to FLT3-ITD mutations could be a new way to detect a significant clinical activity of proteasome inhibitors in AML patients.
Introduction
Acute myeloid leukemia (AML) is a malignant proliferation of myeloid progenitors in bone marrow. Whole-genome sequencing along with RNA and microRNA sequencing and DNA-methylation analysis recently established its molecular map, highlighting the genomic and epigenomic heterogeneity of this malignancy.1 This heterogeneity is one of the crucial challenges for treatment, and optimal therapies would be those that strongly alter crucial regulatory pathways in all AML subtypes.2 Treating leukemia with mutations of the Fms-like tyrosine kinase 3 (FLT3) represents one of these challenges.3 Activating internal tandem duplication (ITD) mutations in the FLT3 gene are found in approximately 30% of patients with AML and are associated with a poor outcome. FTL3-ITD modulates cell proliferation, survival, and differentiation through constitutive activation of canonical pathways, such as PI3K/AKT, STAT5, or MAPK/ERK, and cooperates with other recurrent molecular abnormalities to induce acute leukemia in preclinical models.4-7 However, first generation tyrosine kinase inhibitors (TKIs) targeting FLT3 have proven ineffective.8 Recently, quizartinib, a second generation TKI (formerly AC220) with a higher selectivity for FLT3 and better pharmacologic properties, demonstrated single-agent efficacy by inducing complete responses in patients with refractory disease.9 Interestingly, point mutations at 4 residues within the kinase domain of FLT3-ITD have been detected in patients who developed acquired resistance to quizartinib, with some cases displaying polyclonal resistance, supporting FLT3 as a major therapeutic target in FLT3-ITD-AML.10 Although new TKIs targeting resistance-conferring mutations are currently developed, other therapeutic strategies that modulate the expression of FLT3-ITD are also promising.11 For instance, the transcriptional inhibition of FLT3 via upregulation of miR-29b and disruption of the transcription complex SP1/NFκB(p65) was recently proposed as a new therapeutic strategy for FLT3-ITD AML patients.12 Such strategies would counteract the emergence of subclones with resistance mutations.
The degradation of oncoproteins through induction of autophagy has recently paved the way for new therapeutic options in hematologic malignancies.13 Autophagy is an adaptive and protective cellular program activated under nutrient deprivation, growth factor withdrawal, or metabolic stress to maintain cellular homeostasis and recycle damaged cytoplasmic organelles.14 This dynamic process involves the rearrangement of subcellular membranes to sequester organelles or long-lived proteins, and the subsequent delivery of these contents to lysosomal machinery for degradation and recycling. A set of autophagy genes (ATG) conserved from yeast to mammals encode proteins that are necessary for the coordinated steps of autophagy, such as the formation of autophagosomes, which are double-membrane vesicles. Autophagosomes sequester the cytoplasmic structures targeted for destruction and fuse with lysosome for subsequent degradation. Several signaling pathways including mammalian target of rapamycin complex 1, 5′ AMP-activated protein kinase (AMPK), and PI3-kinase pathways modulate autophagy in response to cellular or environmental stress.15
By exploring the antileukemic activity of proteasome inhibitors in AML, we found that AML samples bearing FLT3-ITD mutations were more sensitive than wild-type samples and that this sensitivity was clearly correlated to the ratio of FLT3-ITD to FLT3 wild-type alleles. Using pharmacologic inhibitors, short hairpin RNA (shRNA) approaches, biochemical and microscopy studies, we demonstrated that proteasome inhibitors induce autophagy in AML cells. Autophagy induction is then responsible for the early degradation of FLT3-ITD, which precedes the inhibition of MAPK/ERK, PI3K/AKT, and STAT5 pathways and subsequent activation of cell death. Furthermore, proteasome inhibitors overcome TKI resistance induced by mutations in the kinase domain of FLT3, suggesting that these compounds may prevent the emergence of mutant clones after TKI treatments in FLT3-ITD AML.
Methods
Cell lines and AML samples
Human myeloid leukemia cell line MV4-11 was purchased from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Leibniz, Germany) and cultured in RPMI containing 10% fetal calf serum. OCI-AML3 cell line was a kind gift form Pierre Brousset Laboratory (Toulouse, France). MOLM-14 cell line was used to generate a FLT3-ITD-D835Y cell line (MOLM-14/TKD). The FLT3-ITD gene was cloned into the pLKO.1-blast lentiviral expression vector (Addgene Plasmid 26655). Mutation producing a D835Y amino-acid substitution within FLT3 kinase domain (FLT3-ITD-D835Y) was generated using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) in accordance with the manufacturer’s instructions using the following (5′-3′) primer: CTTTGGATTGGCTCGATATATCATGAGTGATTCCAAC. We used 293-T packaging cells to produce FLT3-ITD and FLT3-ITD-D835Y lentivirus through cotransfection of FLT3-containing plasmids with lentiviral protein-encoding plasmids. Supernatants were collected over 3 consecutive days beginning 48 hours posttransfection and were stored at −80°C. We plated 106/mL MOLM-14 cells in 100 µL of α-MEM medium and added 5 µL of lentiviral supernatant to the culture. After 3 hours, culture medium was supplemented with 10% fetal bovine serum. Puromycin selection started 48 hours after lentiviral infection and allowed enrichment for FLT3-ITD or FLT3-ITD-D835Y–expressing MOLM-14 cells. AML samples were obtained from patients at the hematology department of Toulouse after consent in accordance with the Declaration of Helsinki. Samples were stored at the Hémopathies Inserm Midi-Pyrénées (HIMIP) collection of Inserm-U1037 (n1DC-2008-307-CPTP1 HIMIP). AML primary samples were cultured in Iscove's modified Dulbecco medium (IMDM) containing 20% fetal calf serum.
Antibodies and reagents
Antibodies antiphospho-Flt-3 (Y591), anti-Lc3-B, anti-caspase 3, antiphospho-Stat-5 (Tyr694), anti-Stat-5, antiphospho-Akt (S473), anti-Akt, antiphospho-p44/42 MAPK Erk1/2 (T202/Y204), and anti-p44/42 MAPK Erk1/2 were obtained from Cell Signaling Technology (Beverly, MA); C-20 anti-Flt-3/Flk-2 was obtained from Santa Cruz Biotechnology (Santa Cruz, CA); mouse monoclonal anti-Flt3 was used for immunofluorescence analysis (MAB812, R&D Systems, Minneapolis, MN); anti-Poly (ADP-ribose) polymerase (PARP) and flow cytometry antibodies anti-hCD45 (YB5.B8), anti-hCD44-PE (4G8), anti-CD33 (HI30), and antimouse IgG1 κ isotype control were obtained from (BD Pharmingen, San Diego, CA). Bortezomib was obtained from Millennium Pharmaceuticals (Cambridge, MA).
Western blot analysis
Proteins were resolved using 4% to 12% polyacrylamide gel electrophoresis Bis-Tris gels (Life Technology, Carlsbad, CA) and electrotransferred to nitrocellulose membranes. After blocking in phosphate-buffered saline (PBS) 0.1% Tween 20% to 5% bovine serum albumin, membranes were immunostained with appropriate antibodies and horseradish peroxidase-conjugated secondary antibodies, and visualized with an enhanced chemoluminescence detection system.
Apoptosis assay
Primary AML samples or cell lines were treated 24 hours with bortezomib, and then 5.105 cells were washed with PBS and resuspended in 300 µL of Annexin-V binding buffer. Annexin-V-FITC and 7-amino-actinomycin D (AAD) (2 µL) were added for 15 minutes at room temperature in the dark. All samples were analyzed by a fluorescence-activated cell sorter (FACS)Calibur flow cytometer (BD Pharmingen, San Diego, CA). For primary samples analysis, leukemic blasts were discriminated against lymphocytes and monocytes with an anti-hCD45 antibody.
Tumor xenograft in NSG or NOD/SCID mice
Xenograft tumors in nonobese diabetic (NOD)/severe combined immunodeficiency (SCID)/IL-2 receptor γ-chain null mice (NSG) were generated by injecting 2.106 MV4-11 into the tail vein. Two weeks after the injection, mice were randomly split in 2 groups and then treated 2 times the first week with 200 µL bortezomib (0.75 mg/kg; n = 10 mice) or PBS (n = 10 mice) once a week then. After 3 daily doses of bortezomib, half of the mice were sacrificed for analysis. Human cell engraftment (hCD33+/hCD44+/hCD45+ cells) in bone marrow was evaluated by flow cytometry (LSRII Fortessa). Cell death was assessed in hCD33+/hCD44+/hCD45+ cells by Annexin-V labeling and flow cytometry analysis. Human cells were also isolated by flow cytometry cell sorting and analyzed by western blot using the appropriate antibodies. For MOLM-14, MOLM-14/TKD, and OCI-AML3 in vivo experiments, 2.106 cells were injected into the tail vein of NSG mice. After 2 weeks, mice were treated twice a week with 200 µL bortezomib (0.75 mg/kg, IP) or PBS until death. For OCI-AML3, human cells were also isolated by flow cytometry cell sorting (hCD33+/hCD44+/hCD45+/Annexin-V−) and analyzed by immunoblotting. Xenograft tumors in nonobese diabetic (NOD)/severe combined immunodeficiency (SCID) mice were generated by injecting subcutaneously 2.106 MV4-11 cells in 100 µL of PBS cells on both flanks. Once the tumors reached 50 to 100 mm3 in size, animals were treated 2 times per week with 200 µL of bortezomib (1 mg/kg, IP) or vehicle (PBS). Tumor dimensions were measured with a caliper on days indicated and volume calculated using the formula: v = A*B2/2, where A is the larger diameter and B is the smaller diameter. All experiments were conducted in accordance with the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care International.
Statistical analysis
Data from 3 independent experiments were reported as mean ± standard error of the mean. Statistical analyses were performed using unpaired two-tailed Student t tests with Prism 5 software (GraphPad Software Inc., La Jolla, CA). A log-rank test was used for the mouse survival analysis and Kaplan-Meier survival curve was used to show the results.
Results
FLT3-ITD sensitizes AML cells to bortezomib treatment
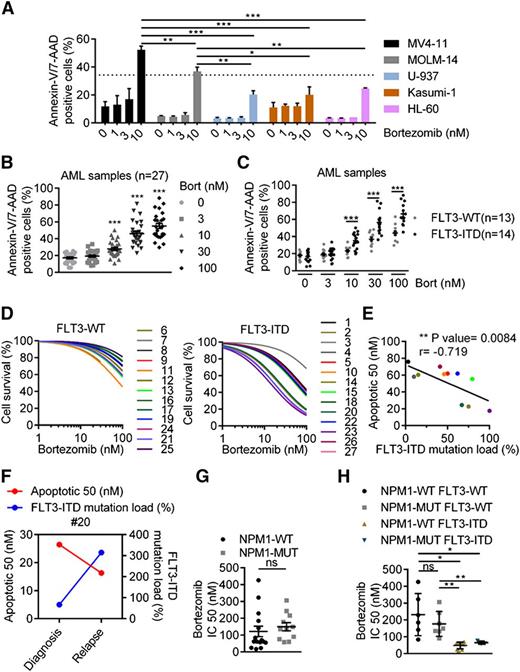
Although in vitro antileukemic activity of proteasome inhibitors has been documented in AML cells, it has not been investigated in the genetic context of this disease.16,17 Thus, we assessed the antileukemic activity of a well-known proteasome inhibitor, bortezomib, according to the FLT3-ITD mutational status, which is the most frequent genetic event in AML.18 We first evaluated the impact of bortezomib exposure on cell proliferation and apoptosis in a panel of leukemic cell lines harboring or not FLT3-ITD mutations. Clinically, achievable concentrations of bortezomib-induced apoptotic cell death in all tested cell lines, but more potently in the FLT3-ITD–positive cell lines, MV4-11, and MOLM-14, compared with the FLT3-wild type cell lines U-937, Kasumi-1, and HL-60 (Figure 1A). Using a blue trypan dye exclusion assay, we further confirmed that bortezomib dramatically abrogated cell survival of MV4-11 and MOLM-14, although it mainly induced a cytostatic effect in U-937 (supplemental Figure 1A). Interestingly, the FLT3-ITD homozygous cell line, MV4-11, was more sensitive than the heterozygous line, MOLM-14, suggesting that the FLT3-ITD mutational load may determine susceptibility to bortezomib-induced cell death (Figure 1A). In a panel of 27 primary AML samples with intermediate-risk cytogenetics, bortezomib-induced apoptosis in a dose-dependent manner (Figure 1B). However, FLT3-ITD AML samples (n = 14) were more sensitive than wild-type samples (n = 13), as the 24 hour IC50 was 4 times lower in mutated compared with wild-type samples (mean IC50 42 nM vs 170 nM) (Figure 1C and supplemental Figure 1B). Similar to the mutational load-based results obtained in FLT3-ITD cell lines, 2 main response patterns were observed in FLT3-ITD patient samples, indicating a heterogeneous response to bortezomib and likely reflecting the heterogeneity of FLT3-ITD mutational load observed in clinic19 (Figure 1D). Indeed, when assessing bortezomib activity, according to the FLT3-ITD mutant to wild-type allele ratio, we found a positive correlation between the IC50 and the FLT3-ITD mutational load, as samples with a higher ratio were the most sensitive (P = .0084; r = −0.719) (Figure 1E). Moreover, the efficacy of bortezomib was also improved at relapse compared with diagnosis in a patient whose mutational load increased with disease progression (Figure 1F). NPM1 mutations are driver mutations frequently associated with FLT3-ITD in cytogenetically normal AML.20 When stratifying samples, according to NPM1 mutational status or to NPM1wt/FLT3wt, NPM1mut/FLT3wt, NPM1wt/FLT3-ITD, and NPM1mut/FLT3-ITD genotypes, the NPM1 mutation did not impact sensitivity to bortezomib. In fact, sensitivity to the drug remained solely dependent on the presence of FLT3-ITD (Figure 1G-H). Altogether, these results suggest that FLT3-ITD sensitizes AML cell lines and primary patient samples to bortezomib-induced cell death.
Antileukemic activity of bortezomib is linked to FLT3-ITD expression. (A) FLT3-ITD AML cell lines are more sensitive than FLT3-WT to bortezomib (Bort)-induced cell death. Apoptosis was evaluated by treating MV4-11, MOLM-14, U-937, HL-60, or Kasumi-1 AML cell lines 24 hours with bortezomib at 1, 3, or 10 nM before Annexin-V/7-AAD staining and flow cytometry analysis. (B-C) Bortezomib targets preferentially FLT3-ITD primary AML samples. Frozen primary AML samples were thawed and treated with bortezomib at 3, 10, 30, or 100 nM for 24 hours. Bortezomib-induced apoptosis was evaluated by labeling cells with Annexin-V/7-AAD via fluorescence-activated cell sorter (FACS) analysis. (D) Viability was assessed by Annexin-V/7-AAD via FACS analysis, and IC50 values were calculated using GraphPad Prism software. (E) Correlation between bortezomib sensitivity and FLT3-ITD mutation load estimated by the level of FLT3-ITD to FLT3 wild-type (WT) allele ratio. (F) Bortezomib activity and FLT3-ITD mutational load in a patient sample assessed at diagnosis and relapse. (G-H). Antileukemic activity of bortezomib, according to NPM1 mutational status. *P < .05; **P < .01; ***P < .001.
Antileukemic activity of bortezomib is linked to FLT3-ITD expression. (A) FLT3-ITD AML cell lines are more sensitive than FLT3-WT to bortezomib (Bort)-induced cell death. Apoptosis was evaluated by treating MV4-11, MOLM-14, U-937, HL-60, or Kasumi-1 AML cell lines 24 hours with bortezomib at 1, 3, or 10 nM before Annexin-V/7-AAD staining and flow cytometry analysis. (B-C) Bortezomib targets preferentially FLT3-ITD primary AML samples. Frozen primary AML samples were thawed and treated with bortezomib at 3, 10, 30, or 100 nM for 24 hours. Bortezomib-induced apoptosis was evaluated by labeling cells with Annexin-V/7-AAD via fluorescence-activated cell sorter (FACS) analysis. (D) Viability was assessed by Annexin-V/7-AAD via FACS analysis, and IC50 values were calculated using GraphPad Prism software. (E) Correlation between bortezomib sensitivity and FLT3-ITD mutation load estimated by the level of FLT3-ITD to FLT3 wild-type (WT) allele ratio. (F) Bortezomib activity and FLT3-ITD mutational load in a patient sample assessed at diagnosis and relapse. (G-H). Antileukemic activity of bortezomib, according to NPM1 mutational status. *P < .05; **P < .01; ***P < .001.
Bortezomib induces FLT3 downregulation that leads to cell death
To explore why bortezomib preferentially targeted FLT3-ITD samples, we first assessed its impact on both FLT3 phosphorylation, indicative of its constitutive activation, and expression, which was previously shown to be negatively regulated by bortezomib at the transcriptional level (supplemental Figure 2A).12 Bortezomib strongly downregulated FLT3 protein expression in MV4-11 and MOLM-14 cell lines (Figure 2A-B). Similar results were obtained with 2 other proteasome inhibitors, MG-132 and carfilzomib (supplemental Figure 2B-C). Pathways that are activated by FLT3-ITD (ie, PI3-K/Akt, STAT5, and MAPK/ERK) were significantly downregulated with bortezomib treatment (Figure 2C). In addition, bortezomib also affected protein expression of the receptor in primary AML samples (Figure 2D). Because many kinases including FLT3-ITD are subject to proteolytic cleavage by caspases, next we sought to determine which event between apoptosis and FLT3-ITD downregulation occurs first.21,22 Kinetic experiments in MV4-11, measuring both apoptosis and FLT3-ITD protein level after exposure to 10 nM bortezomib, showed that the decrease of the receptor preceded apoptosis, which occurred only when FLT3-ITD level was reduced by at least 50% (Figure 2E-F). Furthermore, we also showed that caspase inhibitors (Z-VAD-FMK and Z-DEVD-FMK), although partially inhibited bortezomib-induced apoptosis (supplemental Figure 2D), they completely failed to rescue FLT3-ITD protein expression (supplemental Figure 2E). This data indicates that caspases do not mediate FLT3-ITD downregulation in this model and suggests that it is the loss of FLT3-ITD that initiates cell death.
FLT3-ITD protein expression is downregulated by bortezomib and precedes cell death. (A-B) FLT3-ITD protein level is downregulated by bortezomib (Bort). MV4-11 or MOLM14 or were treated 24 hours with bortezomib at different concentrations. Total cell lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with the appropriate antibodies. (C) MOLM-14 cells were treated 24 hours with 10 nM bortezomib and then analyzed by western blot. (D) FLT3-ITD–positive patient samples were exposed 24 hours with bortezomib at different concentrations before immunoblotting analysis. (E-F) FLT3-ITD downregulation precedes cell death. MV4-11 cells were treated with 10 nM bortezomib for 8 to 24 hours before cell lysis and western blot analysis using the appropriate antibodies. FLT3 protein expression levels were determined by densitometry analysis (Image J software). In parallel, cell death was evaluated by Annexin-V staining and flow cytometry analysis. Cell death (% of maximal effect) represents the ratio ([Annexin-V+ cells t = x*hours/Annexin-V+ cells t = 24 hours] ×100).
FLT3-ITD protein expression is downregulated by bortezomib and precedes cell death. (A-B) FLT3-ITD protein level is downregulated by bortezomib (Bort). MV4-11 or MOLM14 or were treated 24 hours with bortezomib at different concentrations. Total cell lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with the appropriate antibodies. (C) MOLM-14 cells were treated 24 hours with 10 nM bortezomib and then analyzed by western blot. (D) FLT3-ITD–positive patient samples were exposed 24 hours with bortezomib at different concentrations before immunoblotting analysis. (E-F) FLT3-ITD downregulation precedes cell death. MV4-11 cells were treated with 10 nM bortezomib for 8 to 24 hours before cell lysis and western blot analysis using the appropriate antibodies. FLT3 protein expression levels were determined by densitometry analysis (Image J software). In parallel, cell death was evaluated by Annexin-V staining and flow cytometry analysis. Cell death (% of maximal effect) represents the ratio ([Annexin-V+ cells t = x*hours/Annexin-V+ cells t = 24 hours] ×100).
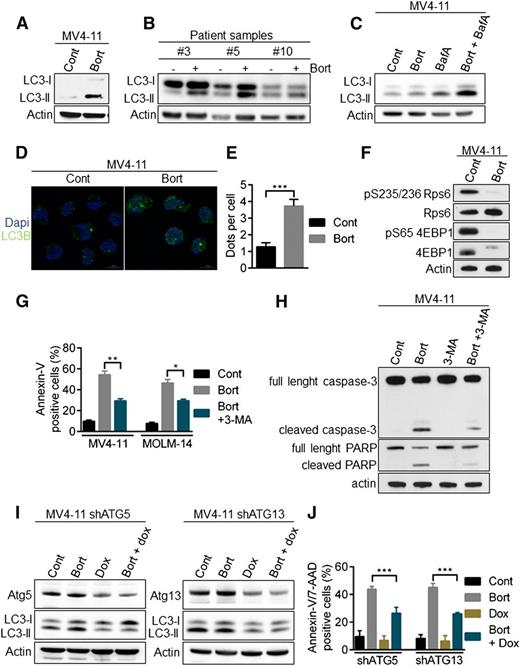
Bortezomib activates cytotoxic autophagy in AML cells
Because caspase-mediated apoptosis did not play a role in FLT3-ITD downregulation in the context of proteasome inhibition, we assessed the role of another major proteolytic pathway, autophagy. Indeed, proteolytic cross-talk between autophagy and the ubiquitin-proteasome pathway exists and previous data have demonstrated that bortezomib could induce apoptosis in AML cells through autophagy.23,24 During autophagy, the microtubule-associated protein 1 light chain 3 (LC3-I) is converted to membrane-bound LC3-II and specifically associates with autophagosomes.25 Treatment of MV4-11 cell line and FLT3-ITD primary samples with bortezomib increased the number of autophagosomes, as indicated by the conversion of LC3-I to LC3-II (Figure 3A-B). The addition of the lysosomal protease inhibitor bafilomycin A to bortezomib treatment further increased the amount of LC3-II, consistent with an active autophagic flux rather than a defect in autophagosome-lysosome fusion (Figure 3C). Moreover, immunofluorescence microscopy analyses also demonstrated an accumulation of LC3-positive structures in MV4-11 cells exposed to bortezomib, indicating an increase in autophagosome formation (Figure 3D-E). As shown by the strong inhibition of RPS6 and 4EBP1 phosphorylation, bortezomib inhibited mammalian target of rapamycin complex 1, one of the best-characterized negative regulators of autophagy (Figure 3F). Taken together, these data show that bortezomib induces autophagy in AML cells.
Activation of autophagy by bortezomib in AML cells. (A-B) LC3-II accumulation after bortezomib (Bort) exposure. MV4-11 or primary AML samples were treated 24 hours with 10 or 30 nM bortezomib, respectively. Total cell lysates were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and immunoblotted with the indicated antibodies. (C) Bortezomib induces an active autophagic flux. MV4-11 cells were incubated 24 hours with 10 nM bortezomib alone or in combination with 10 nM Bafilomycin A for the last 2 hours. Cells were lysed and LC3-I to LC3-II conversion was assessed by western blot analysis using the appropriate antibodies. (D) Immunofluorescence staining of LC3 positive structures. MV4-11 cells were exposed 24 hours with 10 nM bortezomib before immunofluorescence analysis using LC3-B antibody and an antimouse-Alexa 488 antibody, and labeling with 4,6 diamidino-2-phenylindole. Analyses were performed on a Zeiss Apotome microscope. (E) Quantification of LC3-positive dots. At least 100 cells per condition of 3 independent experiments were counted. (F) Inhibition of the mammalian target of rapamycin complex 1 pathway. MV4-11 cells were incubated 12 hours with 10 nM bortezomib before cell lysis and western blot analysis. (G-J) Bortezomib induces a cytotoxic autophagy. (G) MV4-11 or MOLM-14 cells were treated 24 hours with 10 nM bortezomib alone or in combination of 5 mM 3-MA before Annexin-V/7-AAD staining and flow cytometry analysis. (H) MV4-11 cells were incubated 24 hours with bortezomib and/or 5 mM 3-MA before immunoblotting analysis using the appropriate antibodies. (I-J) MV4-11 cells were infected with TRIPZ lentiviral shRNA targeting ATG5 or ATG13 and transduced cells were selected by 1 µg/mL puromycin. First, shRNA were induced by 72 hours 1 µg/mL doxycycline treatment and then next the cells were incubated for 24 hours in the presence of 10 nM bortezomib. (I) Total cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies (J) Apoptosis was evaluated after Annexin-V/7-AAD labeling via flow cytometry. *P < .05; **P < .01; ***P < .001.
Activation of autophagy by bortezomib in AML cells. (A-B) LC3-II accumulation after bortezomib (Bort) exposure. MV4-11 or primary AML samples were treated 24 hours with 10 or 30 nM bortezomib, respectively. Total cell lysates were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and immunoblotted with the indicated antibodies. (C) Bortezomib induces an active autophagic flux. MV4-11 cells were incubated 24 hours with 10 nM bortezomib alone or in combination with 10 nM Bafilomycin A for the last 2 hours. Cells were lysed and LC3-I to LC3-II conversion was assessed by western blot analysis using the appropriate antibodies. (D) Immunofluorescence staining of LC3 positive structures. MV4-11 cells were exposed 24 hours with 10 nM bortezomib before immunofluorescence analysis using LC3-B antibody and an antimouse-Alexa 488 antibody, and labeling with 4,6 diamidino-2-phenylindole. Analyses were performed on a Zeiss Apotome microscope. (E) Quantification of LC3-positive dots. At least 100 cells per condition of 3 independent experiments were counted. (F) Inhibition of the mammalian target of rapamycin complex 1 pathway. MV4-11 cells were incubated 12 hours with 10 nM bortezomib before cell lysis and western blot analysis. (G-J) Bortezomib induces a cytotoxic autophagy. (G) MV4-11 or MOLM-14 cells were treated 24 hours with 10 nM bortezomib alone or in combination of 5 mM 3-MA before Annexin-V/7-AAD staining and flow cytometry analysis. (H) MV4-11 cells were incubated 24 hours with bortezomib and/or 5 mM 3-MA before immunoblotting analysis using the appropriate antibodies. (I-J) MV4-11 cells were infected with TRIPZ lentiviral shRNA targeting ATG5 or ATG13 and transduced cells were selected by 1 µg/mL puromycin. First, shRNA were induced by 72 hours 1 µg/mL doxycycline treatment and then next the cells were incubated for 24 hours in the presence of 10 nM bortezomib. (I) Total cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies (J) Apoptosis was evaluated after Annexin-V/7-AAD labeling via flow cytometry. *P < .05; **P < .01; ***P < .001.
Importantly, inhibiting early steps of autophagy with the type III PI3K inhibitor, 3-methyladenine (3-MA), decreased the emergence of bortezomib-induced Annexin-V/7-AAD–positive cells (Figure 3G), as well as caspase-3 and poly (ADP-ribose) polymerase (PARP) cleavage (Figure 3H), indicating that autophagy is required to activate apoptotic cell death upon bortezomib treatment. To further investigate the role of autophagy in this model, we transduced MV4-11 with lentiviral vectors encoding inducible shRNA against Atg5 or Atg13, 2 key autophagy proteins. Each protein was silenced by shRNAs, and we confirmed that their depletion inhibited bortezomib-induced autophagy as shown by a decrease in LC3-II accumulation (Figure 3I). Moreover, autophagy inhibition significantly reduced cell death induction (Figure 3J). Altogether, these results demonstrated that bortezomib induces autophagy that in turn leads to cell death in AML cells.
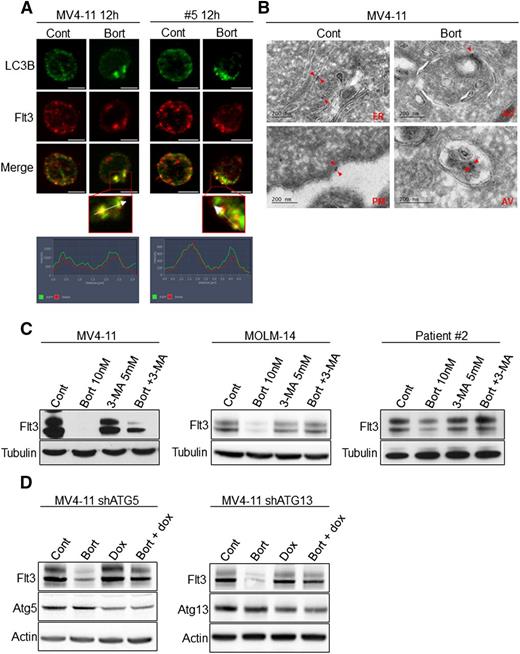
Bortezomib induces the degradation of FLT3-ITD through an autophagy-dependent mechanism
Because autophagy is a catabolic process, next we investigated the role of autophagy in the downregulation of FLT3-ITD after bortezomib treatment. In MV4-11 cells and a primary AML sample, immunofluorescence analyses showed that FLT3-ITD molecules colocalized with LC3-positive structures after treatment with 10 nM bortezomib (Figure 4A). To further study the localization of FLT3-ITD, we performed immunogold labeling coupled with electron microscopy analysis. These studies revealed that immunogold-labeled FLT3-ITD molecules were detectable within autophagosomes after bortezomib treatment indicating that FLT3-ITD could be degraded through bortezomib-induced autophagy (Figure 4B). Indeed, inhibiting autophagy with 3-MA or inducible shRNA against Atg5 or Atg13 restored FLT3-ITD expression in bortezomib-treated cells (Figure 4C-D). We also studied the impact of dynasore, a dynamin inhibitor that participates in autophagosome formation on bortezomib-mediated FLT3-ITD degradation.26 Dynasore also restored FLT3-ITD expression, but importantly also inhibited bortezomib-induced apoptosis in MV4-11 and MOLM-14 cell lines and patient samples (supplemental Figure 3A-D). These results indicate that in AML cells, bortezomib regulates FLT3-ITD expression at the protein level through autophagy induction. It is noteworthy that rapamycin, a potent inducer of autophagy, did not affect FLT3 expression, suggesting that proteasome inhibition is likely required in this mechanism (supplemental Figure 4).
Bortezomib induces FLT3-ITD degradation through autophagy in leukemic cells. (A) Colocalization of FLT3-ITD and LC3-positive structures. MV4-11 cells were treated 12 hours with 10 nM bortezomib (Bort) before immunofluorescence analysis using LC3-B, FLT3, anti-mouse-Alexa 488, anti-rabbit A594 antibodies, and labeling with 4,6 diamidino-2-phenylindole. FLT3-ITD primary AML sample #8 was treated 12 hours with 30 nM bortezomib and analyzed by immunofluorescence with LC3-B and FLT3 antibodies. Analyses were performed on a Zeiss Apotome microscope. (B) Detection of FLT3-ITD in autophagosomes. MV4-11 cells were treated 12 hours with 10 nM bortezomib, fixed, and analyzed by electron microscopy using the Tokuyasu method. FLT3-ITD was labeled using an anti-FLT3 antibody and a secondary antibody coupled to gold beads. Observations were realized on a JEOL JEL-1400 transmission electron microscope. (C-D) Autophagy inhibition rescues FLT3-ITD protein expression. (C) MV4-11, MOLM-14, or patient sample #2 were treated 24 hours with 10 nM or 30 nM bortezomib before cell lysis and immunoblotting analysis using indicated antibodies. (D) Atg5 or Atg13 shRNA were induced by 72 hours treatment with 1 µg/mL doxycycline. Cells were then incubated 24 hours with 10 nM bortezomib and total cell lysates were analyzed by western blot.
Bortezomib induces FLT3-ITD degradation through autophagy in leukemic cells. (A) Colocalization of FLT3-ITD and LC3-positive structures. MV4-11 cells were treated 12 hours with 10 nM bortezomib (Bort) before immunofluorescence analysis using LC3-B, FLT3, anti-mouse-Alexa 488, anti-rabbit A594 antibodies, and labeling with 4,6 diamidino-2-phenylindole. FLT3-ITD primary AML sample #8 was treated 12 hours with 30 nM bortezomib and analyzed by immunofluorescence with LC3-B and FLT3 antibodies. Analyses were performed on a Zeiss Apotome microscope. (B) Detection of FLT3-ITD in autophagosomes. MV4-11 cells were treated 12 hours with 10 nM bortezomib, fixed, and analyzed by electron microscopy using the Tokuyasu method. FLT3-ITD was labeled using an anti-FLT3 antibody and a secondary antibody coupled to gold beads. Observations were realized on a JEOL JEL-1400 transmission electron microscope. (C-D) Autophagy inhibition rescues FLT3-ITD protein expression. (C) MV4-11, MOLM-14, or patient sample #2 were treated 24 hours with 10 nM or 30 nM bortezomib before cell lysis and immunoblotting analysis using indicated antibodies. (D) Atg5 or Atg13 shRNA were induced by 72 hours treatment with 1 µg/mL doxycycline. Cells were then incubated 24 hours with 10 nM bortezomib and total cell lysates were analyzed by western blot.
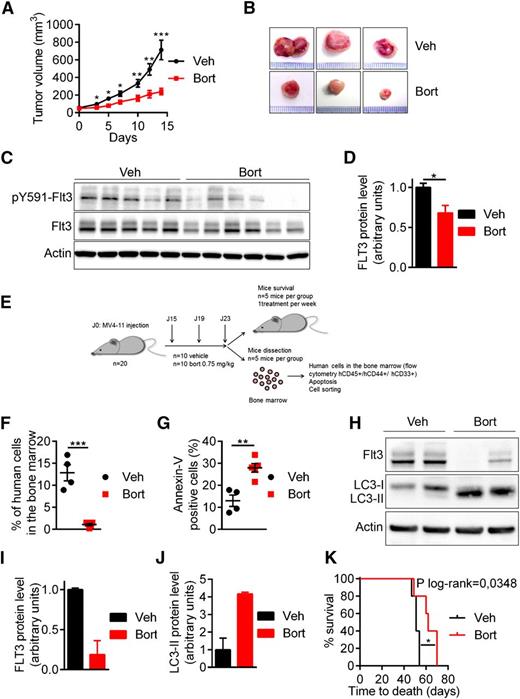
Bortezomib downregulates FLT3-ITD protein expression and inhibits FLT3-ITD driven leukemia in vivo
The activity of bortezomib was addressed in vivo using 2 immunocompromised mouse models engrafted with MV4-11 cells. The first model used was the NOD/SCID xenograft model in which MV4-11 cells were subcutaneously implanted in NOD/SCID mice before bortezomib treatment. As shown in Figure 5A-B, bortezomib significantly inhibited tumor growth and strongly reduced FLT3-ITD protein expression level in treated mice compared with vehicle (Figures 5C-D). In the second model, MV4-11 cells were injected in the tail vein of NSG mice to generate a more relevant physiopathological model. After establishment of the disease, mice were treated with bortezomib at 0.75 mg/kg for 3 injections before processing cells for functional experiments or treating once a week for survival analysis (Figure 5E). Bortezomib strongly inhibited leukemia progression, as indicated by the reduced percentage of human leukemic cells (hCD45+/hCD33+/hCD44+) present in the murine bone marrow (Figure 5F). Furthermore, bortezomib also affected FLT3-ITD protein level in this model (Figure 5H-I) and stimulated the conversion of LC3-I to LC3-II, indicating induction of autophagy in vivo (Figure 5H-J). Features of apoptosis were detected (Figure 5G) and mouse survival was significantly prolonged by bortezomib (P = .0348) (Figure 5K). Thus, these results demonstrated that bortezomib promotes FLT3-ITD degradation and inhibits leukemia progression in vivo.
In vivo antileukemic activity of bortezomib in FLT3-ITD driven leukemia. (A-D) Bortezomib (Bort) inhibited tumor growth and targeted FLT3-ITD protein expression in NOD/SCID mice subcutaneously engrafted with MV4-11 cells. Mice were treated twice a week with 1 mg/mL bortezomib IP. (A-B) Tumor growth was evaluated by measuring tumors with a caliper. (C) Tumor cells were lysed and analyzed by western blot using indicated antibodies. (D) Western blot quantification of FLT3-ITD protein expression. (E-J) Antileukemic activity of bortezomib in NSG mice engrafted by IV injection of MV4-11 cells. After engraftment mice were treated twice a week with IV injections of 0.75 mg/kg bortezomib. After 3 daily doses of bortezomib, vehicle mice (n = 5) and treated mice (n = 5) were sacrificed and analyzed. (F) Percentage of human cells in the murine bone marrow was assessed by hCD45+/hCD33+/hCD44+ labeling and flow cytometry. (G) In vivo bortezomib-induced cell death was evaluated onto the hCD45+/hCD33+/hCD44+ compartment after Annexin-V staining via flow cytometry. (H) Viable human cells hCD33+/hCD45+/Annexin-V− were cell sorted by flow cytometry, lysed, resolved by sodium dodecyl sulfate- polyacrylamide gel electrophoresis, and immunoblotted with indicated antibodies. (I-J) Ratio FLT3-ITD/actin (I) or LC3-II/actin (J) using GeneTool software (western blot quantification). (K) Kaplan Meier survival curves of mice treated by bortezomib or vehicle (Veh). *P < .05; **P < .01; ***P < .001.
In vivo antileukemic activity of bortezomib in FLT3-ITD driven leukemia. (A-D) Bortezomib (Bort) inhibited tumor growth and targeted FLT3-ITD protein expression in NOD/SCID mice subcutaneously engrafted with MV4-11 cells. Mice were treated twice a week with 1 mg/mL bortezomib IP. (A-B) Tumor growth was evaluated by measuring tumors with a caliper. (C) Tumor cells were lysed and analyzed by western blot using indicated antibodies. (D) Western blot quantification of FLT3-ITD protein expression. (E-J) Antileukemic activity of bortezomib in NSG mice engrafted by IV injection of MV4-11 cells. After engraftment mice were treated twice a week with IV injections of 0.75 mg/kg bortezomib. After 3 daily doses of bortezomib, vehicle mice (n = 5) and treated mice (n = 5) were sacrificed and analyzed. (F) Percentage of human cells in the murine bone marrow was assessed by hCD45+/hCD33+/hCD44+ labeling and flow cytometry. (G) In vivo bortezomib-induced cell death was evaluated onto the hCD45+/hCD33+/hCD44+ compartment after Annexin-V staining via flow cytometry. (H) Viable human cells hCD33+/hCD45+/Annexin-V− were cell sorted by flow cytometry, lysed, resolved by sodium dodecyl sulfate- polyacrylamide gel electrophoresis, and immunoblotted with indicated antibodies. (I-J) Ratio FLT3-ITD/actin (I) or LC3-II/actin (J) using GeneTool software (western blot quantification). (K) Kaplan Meier survival curves of mice treated by bortezomib or vehicle (Veh). *P < .05; **P < .01; ***P < .001.
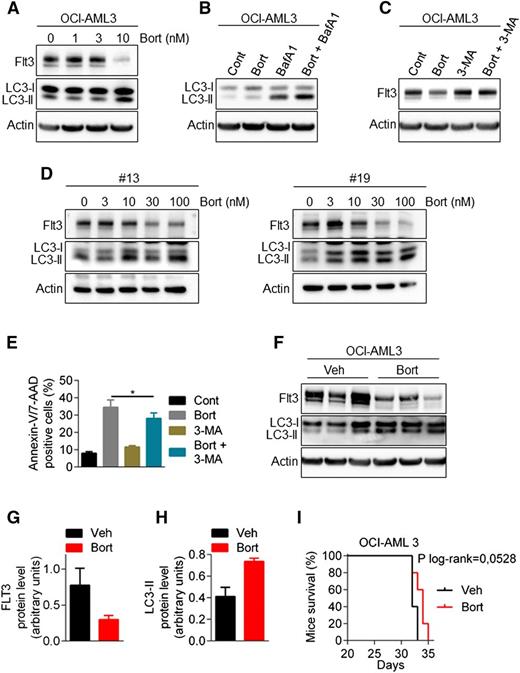
Bortezomib also regulates wild-type FLT3
To determine whether wild-type FLT3 is equally susceptible to degradation by autophagy, we used both the OCI-AML3 cell line (NPM1mut/FLT3wt) and FLT3-wt AML samples. As shown in Figure 6A-E, bortezomib downregulated FLT3 protein expression, stimulated the conversion of LC3-I to LC3-II, and induced an autophagic flux. The 3-MA rescued the expression of FTL3 expression suggesting that similar to FLT3-ITD, the expression of the wild-type receptor is also subjected to autophagic regulation with bortezomib treatment. Furthermore, in vivo experiment using NSG mice injected IV with OCI-AML3 cells showed that bortezomib increased the amount of LC3-II and downregulated FLT3 expression (Figure 6F-H). However, this did not translate into a significant impact on mice survival (Figure 6I).
Bortezomib regulates wild-type FLT3 expression. (A) Downregulation of FLT3-wild-type by bortezomib (Bort). OCI-AML3 cells were treated for 24 hours with increasing concentrations of bortezomib. Total cell lysates were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and immunoblotted with the indicated antibodies. (B) Bortezomib induces an active autophagic flux in FLT3-WT cells. OCI-AML3 cells were incubated for 24 hours with 10 nM bortezomib alone or in combination with 10 nM Bafilomycin A for the last 2 hours. Cells were lysed and LC3-I to LC3-II conversion was assessed by western blot analysis. (C) Inhibition of autophagy rescues bortezomib-induced downregulation of FLT3-WT. OCI-AML3 were treated for 24 hours with 10 nM bortezomib alone or in combination with 5 mM 3-MA, before cell lysis and immunoblotting analysis. (D) Bortezomib effects in primary FLT3-WT AML samples. Patient samples were treated 24 hours with increasing concentrations of bortezomib and then analyzed by western blot. (E) Autophagy inhibition by 3-MA affects bortezomib-induced cell death in OCI-AML3 cells. OCI-AML3 cells were treated 24 hours with 10 nM bortezomib alone or in combination of 5 mM 3-MA before Annexin-V/7-AAD staining and flow cytometry analysis. (F-I) Activity of bortezomib in NSG mice engrafted by IV injection of OCI-AML3 cells. After engraftment, mice were treated twice a week with intraperitoneal injections of 0.75 mg/kg bortezomib. (F) Viable human cells hCD33+/hCD45+/Annexin-V− were cell sorted by flow cytometry, lysed, resolved by SDS-PAGE, and immunoblotted with indicated antibodies. (G-H) Ratio FLT3-ITD/actin (G) or LC3-II/actin (H) using GeneTool software. (I) Kaplan-Meier survival curves of mice treated by bortezomib or vehicle.
Bortezomib regulates wild-type FLT3 expression. (A) Downregulation of FLT3-wild-type by bortezomib (Bort). OCI-AML3 cells were treated for 24 hours with increasing concentrations of bortezomib. Total cell lysates were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and immunoblotted with the indicated antibodies. (B) Bortezomib induces an active autophagic flux in FLT3-WT cells. OCI-AML3 cells were incubated for 24 hours with 10 nM bortezomib alone or in combination with 10 nM Bafilomycin A for the last 2 hours. Cells were lysed and LC3-I to LC3-II conversion was assessed by western blot analysis. (C) Inhibition of autophagy rescues bortezomib-induced downregulation of FLT3-WT. OCI-AML3 were treated for 24 hours with 10 nM bortezomib alone or in combination with 5 mM 3-MA, before cell lysis and immunoblotting analysis. (D) Bortezomib effects in primary FLT3-WT AML samples. Patient samples were treated 24 hours with increasing concentrations of bortezomib and then analyzed by western blot. (E) Autophagy inhibition by 3-MA affects bortezomib-induced cell death in OCI-AML3 cells. OCI-AML3 cells were treated 24 hours with 10 nM bortezomib alone or in combination of 5 mM 3-MA before Annexin-V/7-AAD staining and flow cytometry analysis. (F-I) Activity of bortezomib in NSG mice engrafted by IV injection of OCI-AML3 cells. After engraftment, mice were treated twice a week with intraperitoneal injections of 0.75 mg/kg bortezomib. (F) Viable human cells hCD33+/hCD45+/Annexin-V− were cell sorted by flow cytometry, lysed, resolved by SDS-PAGE, and immunoblotted with indicated antibodies. (G-H) Ratio FLT3-ITD/actin (G) or LC3-II/actin (H) using GeneTool software. (I) Kaplan-Meier survival curves of mice treated by bortezomib or vehicle.
Bortezomib overcomes acquired resistance to quizartinib
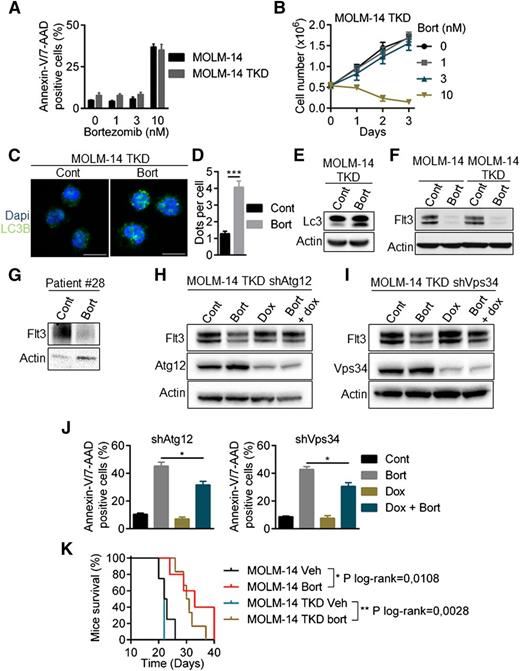
Overcoming resistance to FLT3 inhibitors, including AC220 (quizartinib), is a crucial challenge for AML treatment. Drug-resistant kinase domain mutations have been identified in quizartinib-treated patients. These mutations mainly affect the activation loop at the D835 residue and reduce the binding affinity of the FLT3 inhibitor. We analyzed the sensitivity of a double-mutant MOLM-14/TKD cell line that constitutively expresses both ITD and TKD-D835Y mutations. As shown in supplemental Figure 5A-B, the D835Y mutation conferred a high level of resistance to quizartinib. Conversely, bortezomib had a similar efficacy in both cell lines (Figure 7A-B) and induced autophagy in MOLM-14/TKD cells, as shown by the accumulation of LC3-positive structures (Figure 7C-D) and LC3-I to LC3-II conversion (Figure 7E). Bortezomib downregulated FLT3 expression in MOLM-14/TKD cells and in a patient sample bearing the D835Y mutation (Figure 7F-G). Atg12 and Vps34 depletion using specific shRNAs abrogated this effect, indicating that autophagy also mediated the degradation of FTL3-ITD in quizartinib-resistant cells (Figure 7H-I). In addition, depletion of Atg12 or Vps34 significantly reduced bortezomib-induced cell death (Figure 7J). Altogether, these results show that quizartinib-resistant cells are sensitive to bortezomib through the degradation of FLT3-ITD induced by autophagy. Moreover, bortezomib improved overall survival in mice engrafted with MOLM-14 and MOLM-14/TKD cells (Figure 7K).
Bortezomib overcomes resistance to quizartinib induced by tyrosine kinase domain mutations of FLT3-ITD. (A-B) MOLM-14/TKD-D835Y cells were as sensitive to bortezomib (Bort) as parental MOLM-14. (A) Cells were treated with bortezomib before Annexin-V/7-AAD labeling and analysis by flow cytometry. (B) MOLM-14/TKD cell proliferation upon bortezomib treatment was assessed by trypan blue counting. (C-J) Activation of autophagy by bortezomib in MOLM-14/TKD cells. (C) MOLM-14/TKD cells were incubated 24 hours with 10 nM bortezomib before immunofluorescence analysis using LC3-B antibody and an anti-mouse-Alexa 488 antibody, and labeling with DAPI. Analyses were performed on a Zeiss LSM710 confocal microscope. (D) Quantification of LC3-positive dots. At least 100 cells per condition of three independent experiments were counted. (E) Cells were treated 24 hours with 10 nM bortezomib before immunoblotting analyses using appropriate antibodies. (F-G) Downregulation of FLT3-ITD after bortezomib treatment. MOLM-14, MOLM-14/TKD cells (F) or a FLT3-TKD primary AML sample (G) were treated 24 hours with 10 nM or 3 nM bortezomib, respectively, and then analyzed by western-blotting. (H-I) MOLM-14/TKD cells were infected with TRIPZ lentiviral shRNA targeting ATG12 or VPS34. ShRNA expressions were induced by 72 hours 1 µg/mL doxycycline treatment and cells were incubated 24 hours with 10 nM bortezomib. Cells were then lysed and analyzed by immunoblotting. (J) Cells were treated 24 hours with 10 nM bortezomib in the presence or not of doxycycline and apoptosis was assessed by flow cytometry after Annexin-V/7-AAD staining. (K) Kaplan Meier survival curves of MOLM-14 and MOLM-14/TKD mice treated by bortezomib or vehicle. Dapi, 4,6 diamidino-2-phenylindole. *P < .05; **P < .01; ***P < .001.
Bortezomib overcomes resistance to quizartinib induced by tyrosine kinase domain mutations of FLT3-ITD. (A-B) MOLM-14/TKD-D835Y cells were as sensitive to bortezomib (Bort) as parental MOLM-14. (A) Cells were treated with bortezomib before Annexin-V/7-AAD labeling and analysis by flow cytometry. (B) MOLM-14/TKD cell proliferation upon bortezomib treatment was assessed by trypan blue counting. (C-J) Activation of autophagy by bortezomib in MOLM-14/TKD cells. (C) MOLM-14/TKD cells were incubated 24 hours with 10 nM bortezomib before immunofluorescence analysis using LC3-B antibody and an anti-mouse-Alexa 488 antibody, and labeling with DAPI. Analyses were performed on a Zeiss LSM710 confocal microscope. (D) Quantification of LC3-positive dots. At least 100 cells per condition of three independent experiments were counted. (E) Cells were treated 24 hours with 10 nM bortezomib before immunoblotting analyses using appropriate antibodies. (F-G) Downregulation of FLT3-ITD after bortezomib treatment. MOLM-14, MOLM-14/TKD cells (F) or a FLT3-TKD primary AML sample (G) were treated 24 hours with 10 nM or 3 nM bortezomib, respectively, and then analyzed by western-blotting. (H-I) MOLM-14/TKD cells were infected with TRIPZ lentiviral shRNA targeting ATG12 or VPS34. ShRNA expressions were induced by 72 hours 1 µg/mL doxycycline treatment and cells were incubated 24 hours with 10 nM bortezomib. Cells were then lysed and analyzed by immunoblotting. (J) Cells were treated 24 hours with 10 nM bortezomib in the presence or not of doxycycline and apoptosis was assessed by flow cytometry after Annexin-V/7-AAD staining. (K) Kaplan Meier survival curves of MOLM-14 and MOLM-14/TKD mice treated by bortezomib or vehicle. Dapi, 4,6 diamidino-2-phenylindole. *P < .05; **P < .01; ***P < .001.
Discussion
In this study, we have demonstrated that proteasome inhibitors regulate the expression of FLT3-ITD at the protein level through induction of autophagy, which abrogates FLT3-ITD signaling and induces cell death. Although previous studies determined that bortezomib can interfere with the transcription of FLT3-ITD, our results uncover a second level of regulation of this oncoprotein and provide more detail as to why FLT3-ITD AML cells are particularly sensitive to proteasome inhibitors.12,27
Defects in autophagy are associated with various diseases including cancer.28 The role of autophagy in cancer, however, is complex and depends on tumor subtypes, tumor progression stages, cellular context, or drugs that induce this process.29 For example, although autophagy has a demonstrated role in tumor suppression, other data suggest that autophagy has the capacity to confer a survival advantage in established tumors, particularly under chemotherapeutic stress.30 Autophagy may also promote cell death in apoptosis-resistant tumor cells. Together, these results demonstrate the need to fully explore the role of autophagy according to cancer type, including hematologic malignancies.13 For instance, in myeloid leukemia, autophagy induced by arsenic trioxide or all-trans retinoic acid contributes to cell death through the degradation of fusion oncoproteins such as promyelocytic leukemia–retinoic acid receptor alpha (PML-RARA) in acute promyelocytic leukemia cells or breakpoint cluster region–Abelson (BCR-ABL) in chronic myelocytic leukemia cells, whereas it does not mediate degradation of AML1-ETO (another oncoprotein involved in AML), and protects AML1-ETO AML cells from cell death induced by histone deacetylase inhibitors.31-34 Thus, elucidating the role of autophagy in genetically defined AML subtypes is critical before exploiting autophagic properties as therapeutic approaches.13 In addition to PML-RARA and BCR-ABL, our study shows that FLT3-ITD represents another leukemogenic protein targeted by autophagy. Whether other candidates could be degraded through this mechanism and offer a therapeutic window remains to be determined. Of note, it has been shown previously that bortezomib triggered a clathrin-mediated endocytosis and lysosomal degradation of C-KIT in AML1-ETO AML cells.35 However, the cellular context, as well as compounds that modulate autophagy, will have to be strictly characterized before choosing a strategy targeting the inhibition or induction of autophagy. For instance, although blocking autophagy with chloroquine has been shown to improve the activity of BCR-ABL inhibitors in CML cells through the eradication of CML stem cells, inducing autophagy with arsenic trioxide led to cell death in the same model through BCR-ABL degradation.31,36 In the FLT3-ITD model, using pharmacologic and genetic approaches, we demonstrated that the induction of autophagy by bortezomib is toxic for leukemic cells. However, it is also plausible that the concomitant inhibition of the ubiquitin-proteasome pathway (UPS) may have played a role. Indeed, histone deacetylase inhibitors has been shown recently to induce proteasomal degradation of FLT3-ITD catalyzed by the E2 ubiquitin conjugase UBCH8 and the E3 ubiquitin ligase SIAH1 in MV4-11 cells.37 As UPS and autophagy are functionally interrelated catabolic processes sharing certain substrates, enhanced degradation by autophagy may have become critical for FLT3-ITD expression under conditions in which UPS was compromised by proteasome inhibitors.38,39 Further studies are needed to clarify which molecular players are involved in this proteasome-to-autophagy switch in the context of FLT3-ITD AML cells.40
Bortezomib has been previously tested for its clinical activity in AML patients as a single agent in early phase studies or in combination with chemotherapy.41,42 As a single agent, bortezomib provided limited clinical activity. However, to our knowledge, no study has analyzed its impact according to FLT3 mutational status. Our results strongly argue for the assessment of proteasome inhibitor efficacy in patients with FLT3-ITD AML, particularly those displaying higher FLT3-ITD allelic ratios or acquired resistance to FLT3 inhibitors, such as quizartinib. The resistance induced by mutations in the tyrosine kinase domain will undoubtedly become a major challenge in these patients and strategies aiming at preventing or overcoming it will be needed.10 Furthermore, poor pharmacologic properties of proteasome inhibitors could also explain the limited activity of bortezomib observed in AML patients. Indeed, it has been recently suggested that a liposomal formulation of bortezomib leads to higher concentrations of the molecule in the bone marrow as compared with free bortezomib, and effectively eradicates AML cells in vivo in an MllPTD/wt:Flt3ITD/wt murine model.27
The results presented here provide a mechanism-based rationale for the study of proteasome inhibitors in AML with FLT3-ITD mutations, which constitute a large subset of patients sharing a high risk of relapse and resistance to conventional chemotherapy and tyrosine kinase inhibitors. Those patients with a high FLT3-ITD mutant to wild-type allele ratio are expected to be particularly sensitive to proteasome inhibitors and such drugs could thus potentially improve outcomes in this genetic subtype of AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We acknowledge all the members of the Gaël Adolescent Espoir Leucémie association, Valérie Duplan-Eche, and Fatima L’Faqihi-Olive for technical assistance at the flow cytometry core facility of INSERM UMR1043. We also thank Sarah Scotland for the corrections of the manuscript.
This work was supported by the French government grant under the “Investissement d’avenir” program (ANR-11-PHUC-001), the Institut National du Cancer (INCA-PLBIO 2012-105), the InnaBioSanté foundation (RESISTAML project), and the La Ligue Contre le Cancer.
Authorship
Contribution: C.L., J.E.S., and C.R. designed the study; C.L., J.E.S., and C.R. interpreted data; E.S., H.B., F.V., M.D, M.-A.H., J.T. performed experiments; F.V., M.D, M.-A.H., J.T., C.L., E.D., and J.E.S. analyzed data; C.J. contributed to writing the manuscript; E.D. performed molecular analyses; S.M. contributed to analyzing and discussing data; C.R. supervised the study; and C.L. and C.R. wrote the manuscript.
Conflict-of-interest disclosure: C.R. has received commercial research grants from Celgene and Chugai and is a consultant/advisory board member of Celgene and Sunesis Laboratories. J.E.S. has received commercial research grant from GSK Laboratory. The remaining authors declare no competing financial interests.
Correspondence: Christian Récher, Service d’Hématologie, Institut Universitaire du Cancer de Toulouse Oncopole, 1 Ave Irène Joliot-Curie, 31059 Toulouse, Cedex 9, France; e-mail: recher.christian@iuct-oncopole.fr.
References
Author notes
J.E.S. and C.R. contributed equally to this study.

![Figure 2. FLT3-ITD protein expression is downregulated by bortezomib and precedes cell death. (A-B) FLT3-ITD protein level is downregulated by bortezomib (Bort). MV4-11 or MOLM14 or were treated 24 hours with bortezomib at different concentrations. Total cell lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with the appropriate antibodies. (C) MOLM-14 cells were treated 24 hours with 10 nM bortezomib and then analyzed by western blot. (D) FLT3-ITD–positive patient samples were exposed 24 hours with bortezomib at different concentrations before immunoblotting analysis. (E-F) FLT3-ITD downregulation precedes cell death. MV4-11 cells were treated with 10 nM bortezomib for 8 to 24 hours before cell lysis and western blot analysis using the appropriate antibodies. FLT3 protein expression levels were determined by densitometry analysis (Image J software). In parallel, cell death was evaluated by Annexin-V staining and flow cytometry analysis. Cell death (% of maximal effect) represents the ratio ([Annexin-V+ cells t = x*hours/Annexin-V+ cells t = 24 hours] ×100).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/7/10.1182_blood-2015-05-646497/4/m_882f2.jpeg?Expires=1769863289&Signature=g6ABBdyb5B4p5QyQUPrhXo8CmZ1iZX3s5a38~F1S7sosV2QT4UlYdCaeMktrVfPDm5Fh6mK35eZ7EtUnYf-E2KRgZt-r7BfYY24vHU5xtdPPNnFgyZdAvwxVT7f6ZS4RBxsS9c1y57VhSBmmPQNE4uYejkizCaeygp4jvBpjAK50TZTQOKlSnye11O5q9HZGIPNIXhdWdU7gHJ~mDuCen6RpkucPAcTeACurwzL5t72UARbQf6DuoBeGBgmR9e-yJnbesIFbLuhMBCJ0cSYUWET2dt1rapn-Jw7JKOntXUsrdXNAxHE6EAqRiY3vSQf~Aunv25IlBmdZ0WXzr4Wdhg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)