In this issue of Blood, Vallois et al show that genome and transcriptome analyses focused on T-cell receptor (TCR)-related genes can identify new druggable targets particularly valuable in peripheral T-cell lymphomas (PTCLs), in which the pathogenesis is poorly understood and treatment outcome is unfavorable.1
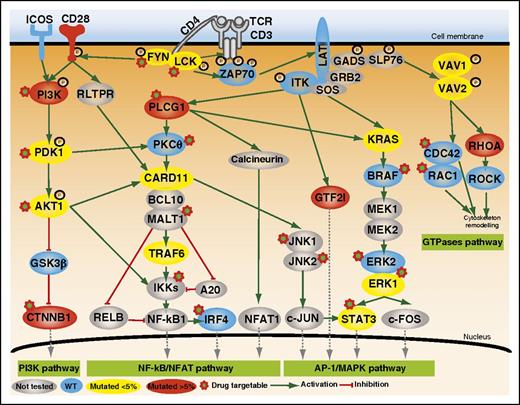
Mutated genes involved in TCR costimulation or intracellular signaling. Positive and inhibitory interactions are depicted as solid green and red arrows, respectively. The protein symbols of mutated genes appear inside colored ovals depending on the frequency of mutation (yellow, <5%; red, >5%). Those of wild-type (WT) genes appear in blue ovals, and not-sequenced genes are in gray. PI3K, phosphatidylinositol 3-kinase. See Figure 2 in the article by Vallois et al that begins on page 1490.
Mutated genes involved in TCR costimulation or intracellular signaling. Positive and inhibitory interactions are depicted as solid green and red arrows, respectively. The protein symbols of mutated genes appear inside colored ovals depending on the frequency of mutation (yellow, <5%; red, >5%). Those of wild-type (WT) genes appear in blue ovals, and not-sequenced genes are in gray. PI3K, phosphatidylinositol 3-kinase. See Figure 2 in the article by Vallois et al that begins on page 1490.
Nodal lymphomas with a T-follicular helper (TFH) immunophenotype include angioimmunoblastic T-cell lymphomas (AITL) and a subset of PTCL not otherwise specified. Recent advances in this field have led the World Health Organization to revise the entity of both nodal and extranodal T-cell and natural killer cell neoplasms, proposing some revisions and new provisional entities.2 Clinically, PTCLs treated with conventional therapies have a poor clinical outcome. Although stem cell transplantation is effective in prolonging disease-free survival in young patients, early disease progression makes even this option difficult.3
Several lines of evidence had previously suggested that TCR activation might be involved in PTCL pathogenesis. For example, the ITK-SYK fusion found in PTCLs mimics the TCR-triggering antigen-independent activation of the TCR-signaling pathway. Therefore, based on multiple studies suggesting that this pathway can induce a T-cell lymphoproliferative disorder, the authors focused on investigating new putative mutations in the costimulatory/TCR cascade working on the hypothesis that abnormal TCR-signaling costimulation of signaling may be relevant to PTCL pathogenesis. In this way, they first applied whole-exome/genome sequencing in a limited discovery cohort. Then, they designed an elegant targeted deep-sequencing approach, consisting of a panel enriched in TCR-signaling elements, which allowed them to capture a broader spectrum of mutations than those detectable using Sanger sequencing and which facilitated finding more information about the frequency of mutated alleles in an extended cohort. Besides RHOA mutations, which had been previously described in 70% of AITL and TFH-like PTCLs,4 they discovered that nearly half of the patients (49%) in this extended cohort harbored mutually exclusive mutations in other TCR-related genes in particular PLCG1 (14.1%), CD28 (9.4%, mutated exclusively in AITL), PI3K elements (7%), CTNNB1 (6%), and GTF21 (6%) (see figure).
To evaluate the importance of these mutations, they functionally assessed mutations in the RHOA gene by pulldown experiments and luciferase reporter assays. Additionally, PLCG1 and CARD11 variants were functionally analyzed by their ability to induce MALT1 protease activity and were tested in luciferase-based assays. Interestingly, the results of these functional analyses revealed that most mutations could be classified as gain-of-function activating mutations. Indeed, the integration of mutational analyses with gene expression profiles allowed them to verify that the TCR-mutated samples exhibited enrichment for molecular signatures reflecting T-cell activation and proliferation.
How do these mutations evolve over the course of disease and in response to chemotherapy? Direct comparisons between samples at diagnosis and at relapses could shed considerable light on the impact of these mutations in lymphoma progression and clinical outcome in the context of intratumor heterogeneity. Therefore, the authors performed preliminary analyses of paired samples in 4 relapsed patients and concluded that the mutational heterogeneity that characterizes AITL samples at diagnosis is essentially conserved after treatment, although 2 patients exhibited 1 additional mutation at relapse involving CTNNB1 and VAV1 genes. Because these analyses were done in a very limited number of sample pairs, additional efforts, by comparing data from targeted gene sequencing and patterns of gene expression in a larger number of paired samples, would be instructive. In this context, it would be reasonable to think that tumors exhibiting intratumor heterogeneity at diagnosis might be genetically different in relapsed samples due to selection of cell clones enriched in driver genes responsible for resistance.5
Another interesting area needing exploration is epigenetics. Because highly recurrent mutations in PTCLs (including AITLs) involve epigenetic modifiers as TET2, DNMT3A, and IDH2,1,4 mapping and analysis of the chromatin state could be a powerful means to detect additional changes of regulatory sequences and activity, which critically contribute to the progression of these lymphomas.6
The authors have integrated genome data into a clinically useful model to conclude that there are no significant differences in the main clinical characteristics or overall survival between TCR-mutated and non-TCR-mutated patients. However, TCR-mutated patients receiving conventional chemotherapy had an increased risk of early progression compared with nonmutated patients. Therefore, it seems clear that using drugs capable of reducing TCR activation, as suggested in this article, may be a very useful choice for treating these types of T-cell lymphomas.
Conflict-of-interest disclosure: The author declares no competing financial interests.