Abstract
Introduction
Adult T-cell leukemia/lymphoma (ATL) is a rare, aggressive disease caused by human T cell lymphotropic virus type 1 (HTLV1) that predominantly affects Japanese and Caribbean populations. (Goncalves, Proietti et al. 2010) Most studies have focused on Japanese cohorts. A recent whole genome/exome sequencing of 81 Japanese ATL cases identified alterations that overlap with the HTLV1 Tax interactome and are highly enriched for T cell receptor-NF-kB signaling, and T cell trafficking (Kataoka, Nagata et al. 2015). We recently showed that ATL in Caribbean patients has a poor prognosis and may have distinctive clinical features when compared to the Japanese cohort (Zell, Assal et al. 2016). In order to determine whether there are genomic differences between the two cohorts, we sequenced 15 ATL samples from the Caribbean cohort.
Methods
In this prospective study we perform comprehensive Next Generation Sequencing to assess the mutational spectrum of ATL in Caribbean patients. Samples were analyzed by complete genetic profiling with the Genoptix Nexcourse complete assay that contains 173 cancer related genes. Patient genomic DNA was utilized to identify relevant single nucleotide variants, insertion/deletions, copy number variations, and translocation fusion genomic alterations in a panel of up to 173 reportable genes at an average mean sequencing depth of 500X coverage.
Results
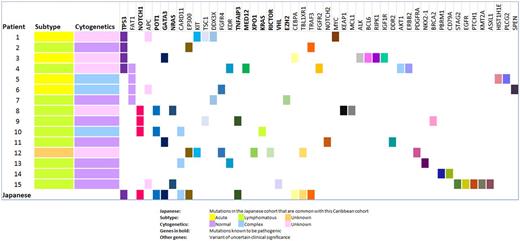
In the 15 patients, a total of 49 genes were found to be mutated ranging from 3 to 7 mutated genes in each patient. The percent of mutated genes amongst the total analyzed ranged from 1.7-4% for each patient and the percent of mutated genes known to be pathogenic ranged from 0-2.3% for each patient. There were 18 known pathogenic mutations in a total of 70 genetic mutations identified in this cohort (25.7%). The mean number of mutations in the acute and lymphomatous subtype was approximately 0.9%.
Genes most commonly mutated in out cohort were TP53 (26.7%), FAT1 (26.7%), NOTCH1 (20%) and APC (20%). Novel mutations not reported by the Japanese cohort but present in our cohort are - NRAS, KRAS, EZH2, RICTOR, XPO1, VHL (known to be pathogenic), FAT1, APC, DDX3X, KIT, NOTCH2, MYC, TSC1, PLCG2, FGFR2, FGFR4, PDGFRA, EGFR, KEAP1, MCL1, ALK, BCL6, RIPK1, IGF1R, DDR2, KDR, AKT1, ERBB2, NKX2-1, BRCA2, PBRM1, CD79A, STAG2, PTCH1, KMT2A, HIST1HE, SPEN, ASXL1 (not known to be pathogenic). Mutations common to both the Japanese and our Caribbean cohort are - CARD11, TNFAIP3, TRAF3 (TCR signaling/NFkB pathway), NOTCH1 (signaling pathway), GATA3, CEBPA, TBL1XR1 (transcriptional regulation), EP300 (epigenetic regulation), TP53, POT1 (Telomere regulation and DNA repair).
Serial analysis of a patient revealed increase in the size of the pathogenic clone with XPO1 mutation. XPO1 has been involved in nuclear transport of p53 and the increase in its clone size occurred with relapse after cytotoxic chemotherapy.
The overall survival (OS) was shorter for TP53 mutated patients (99.3 days) versus non TP53 mutated patients (307.8 days).
Discussion
We report on mutational profiling in the largest cohort of Caribbean ATL and an additional 8 patients will be included in the analysis to be presented at ASH. A significant number of genes found to be mutated affected known pathways in the pathogenesis of ATL. There are known genes which are common between the Japanese and Caribbean cohorts but there are some notable differences.
Previously we showed that the Caribbean cohort has a poorer OS than the Japanese cohort. TP53 mutations are more frequent and associated with decreased OS in this cohort. Genes involved in Wnt pathway - FAT1, APC, NOTCH1 were more frequently mutated in our cohort. FAT1 mutations have not been described in ATL. However, they have been implicated in oncogenesis in solid tumors, chronic lymphocytic leukemia and T-cell acute lymphoblastic leukemia. FAT1 acts as a tumor suppressor gene and encodes a cadherin-like protein, which potently suppresses cancer cell growth. Inactivation of FAT1 via mutation therefore promotes Wnt signaling and tumorigenesis and affects patient survival (Morris, Kaufman et al. 2013). In in-vitro models of ATL, Wnt pathway dysregulation has been shown to promote ATL tumorigenesis (Bellon, Ko et al. 2013, Ma, Yasunaga et al. 2013). Based on this study the driver mutations of the Caribbean cohort appear to be the Wnt signaling pathway which is different from the TCR- NF-kB signaling pathway seen in the Japanese cohort.
Hall:Genoptix, a Novartis Company: Employment.
Author notes
Asterisk with author names denotes non-ASH members.