Abstract
Introduction: Heparanase (HPSE) is an endo-glucuronidase degrading heparan sulfate and playing important roles in angiogenesis, tumor metastasis, inflammation and autoimmunity. The enzyme also regulates histone methylation by binding to target gene regulatory regions. Our previous studies indicated that HPSE gene SNP rs4693608 significantly correlated with the expression level of heparanase among healthy persons. Furthermore, the rs4693608 SNP showed significant association with i) the risk of acute and extensive chronic GVHD, ii) HPSE gene expression both before and after chemotherapy administered as pre-hematopoietic stem cell transplantation conditioning, iii) the time of neutrophils and platelets recovery and iv) HPSE gene expression after lipopolysaccharide (LPS) treatment. Other studies revealed correlations between rs4693608 and sinusoid obstruction syndrome, and with bone morbidity and outcome of multiple myeloma.
Methods: In the present study we cloned 440bp fragment of the HPSE gene (intron 2) that includes rs4693608 SNP using Pgl4.26 minimal promoter vector. Reporter activity of luciferase was detected in U937, KG1, K562, RPMI8226, and CAG cell lines. In addition, nuclear protein extracts from a broad range of malignant cell lines as well as from primary samples of patients with acute myeloid (AML) and lymphatic (ALL) leukemia and normal healthy individuals were subjected to binding to 30bp oligonucleotides probe representing the enhancer region of either the A allele or the G allele of rs4693608 SNP by EMSA (electrophoretic mobility shift assay).
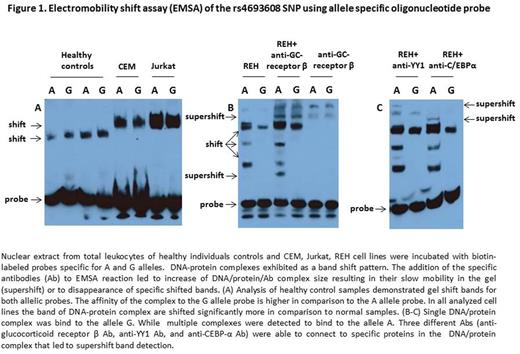
Results:Our results indicate that the 440bp fragment of the HPSE gene exhibits enhancer activity in both the sense and antisense directions of the HPSE gene. Relative luciferase activity was higher in constructs with antisense direction of the enhancer. Moreover, constructs that included the allele G revealed elevated level of luciferase activity in comparison to constructs that included the A allele. Analysis of healthy control samples using EMSA identified gel shift bands for both allelic probes. However, the affinity of the complex to the G allele probe was significantly higher in comparison to the A allele probe (Fig.1A). The DNA-protein complexes were shifted significantly more in malignant cell lines and primary leukemic samples as compared to normal control samples (Fig.1A). Moreover, there were additional nuclear-protein complexes that bind to the A allele probe (Fig.1B). Notably, at least 5 different DNA-protein complexes connected to allele A were found. In order to identify them we used bioinformatics analysis to predict proteins in the DNA/protein complex, followed by super-shift assay. At least three proteins were identified: β-glucocorticoid receptor, Yin Yang1 (YY1) transcription factor, and CCAAT/enhancer-binding protein-α (C/EBPα), which are part of this structured nuclear protein-binding complex (Fig.1B-C).
Conclusions:The study demonstrated that HPSE rs4693608 SNP is located in a strong enhancer. A to G alteration leads to a different enhancer activity as a result of unique DNA-protein complex which binds to DNA regulatory elements in the enhancer. The composition of the DNA-protein complex which binds to the G allele, shifted similarly in all cell lines and primary leukemic samples but not in normal samples. At least 5 different variations of DNA-protein complexes connected to allele A were found, including β-glucocorticoid receptor, YY1 and C/EBPα. Precise identification of the specific proteins that bind to the unique enhancer region will allow modification of the enhancer activity that accelerates the expression of HPSE and thereby influences downstream processes.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.