Abstract
Background
Whole genome, whole exome, and targeted sequencing have identified various genomic alterations in mantle cell lymphoma (MCL). There remains limited data correlating genomic alterations with clinical outcome in MCL. We describe genomic alterations in MCL using a targeted sequencing platform and correlate these alterations with clinical outcome.
Methods
Genomic DNA was isolated from formalin-fixed paraffin-embedded tissue specimens from 55 cases of MCL and screened for clinically relevant mutations by deep-coverage targeted sequencing. Adaptor ligated sequencing libraries were captured by solution hybridization using two custom bait sets targeting 585 biologically significant cancer-related genes for DNA-Seq. Captured libraries were sequenced to high depth (Illumina HiSeq), averaging >300x. Twenty-eight tumor specimens had corresponding matched normal specimens that facilitated identification of somatic mutations with a high degree of confidence. For the remaining 27 unmatched cases, somatic mutations were inferred according to annotated-based rules. Briefly, non-synonymous variants were considered somatic if they met any of the following criteria: 1) previously reported in the COSMIC database at least 5 times or more, 2) previously reported in published MCL genomic data, or 3) truncating mutations including nonsense mutations, frameshift mutations and splicing mutations that are not at the 3' end of the genes. The sequencing data were also used for detecting copy number changes including amplifications of established oncogenes or homozygous deletions of known tumor suppressor genes. Overall survival (OS) analyses were performed using Kaplan-Meier and log-rank tests.
Results
55 cases of newly diagnosed MCL treated at MSKCC from 1999 to 2006 with available specimens were analyzed. Median age of diagnosis was 59 years (range 24-79) with a male predominance (76%). Advanced disease (III/IV) was seen in 93% of cases and an elevated Ki-67 (>=30%) was noted in 41% of patients. Initial treatment included intensive therapy (chemotherapy + ASCT/Allo)(55%), radioimmunotherapy (131I-tositumomab followed by chemotherapy) (20%),bendamustine and an anti-CD20 monoclonal antibody (rituximab or ofatumumab) (11%), single agent anti-CD20 monoclonal antibody (5%), initial observation (7%), and other (2%).
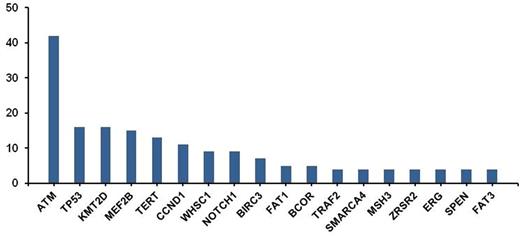
The most frequent alterations, above a level of 4%, are shown in Figure 1. The most commonly altered gene was ATM, a tumor suppressor involved in DNA damage repair, and observed in 42% of cases. In addition, known driver mutations in MCL were found, such as TP53 (16%) and CCND1 (11%). Also an amplification of CCND1 was observed in one patient. Alterations in chromatin modifying genes (e.g. KMT2D, MEF2B, WHSC1, SMARCA4) were common, occurring collectively in 38% of the cases. Recurrent alterations in NOTCH1 were identified in 9% of cases. Alterations in BIRC3 (7%) and TRAF2 (4%), key components of the alternative NF-ĸB pathway and mediators of apoptosis, were identified. Homozygous deletion of BIRC3 was also noted in one patient. Homozygous deletion of CDNK2A was noted in 9% of cases. Recurrent promoter mutations were observed in TERT (Telomerase Reverse Transcriptase) gene (13%), which encodes an enzyme involved in cell immortality.
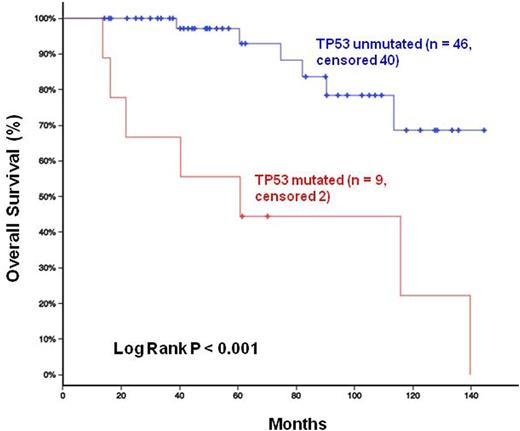
With a median follow-up of 7 years, the presence of TP53 mutations was significantly associated with inferior survival compared to TP53-unmutated patients (P<0.001), Figure 2. Notably, homozygous deletion of CDNK2A and mutations in NOTCH1 and KMT2D were not prognostically significant in our series.
Conclusion:
This analysis further describes the mutational landscape in MCL. Our results are congruent with prior data demonstrating the prognostic relevance of p53 mutations. Several of the genomic alterations identified may have therapeutic significance. For example, TRAF2 and BIRC3 mutations, in the alternative NK-κB pathway, are hypothesized to be associated with ibrutinib resistance. Loss of CDNK2A may confer sensitivity to PRMT5 inhibition. In future,we plan to analyze additional cases of MCL to elucidate the biologic and clinical significance of the observed genetic alterations with the aim of developing mechanism-based, biologically-targeted therapy in MCL.
Kumar:Celgene: Honoraria, Other: Scientific Advisory Board; Celgene: Research Funding; Adaptive Biotechnologies: Research Funding; Seattle Genetics: Research Funding; Pharmacyclics: Research Funding. Zelenetz:Gilead Sciences: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.