Key Points
Isolation and characterization of a high-affinity TCR targeting the intracellular B cell–specific transcription factor BOB1.
T cells expressing a BOB1-specific TCR lysed and eradicated primary multiple myeloma and other B-cell malignancies in vitro and in vivo.
Abstract
Immunotherapy for hematological malignancies or solid tumors by administration of monoclonal antibodies or T cells engineered to express chimeric antigen receptors or T-cell receptors (TCRs) has demonstrated clinical efficacy. However, antigen-loss tumor escape variants and the absence of currently targeted antigens on several malignancies hamper the widespread application of immunotherapy. We have isolated a TCR targeting a peptide of the intracellular B cell–specific transcription factor BOB1 presented in the context of HLA-B*07:02. TCR gene transfer installed BOB1 specificity and reactivity onto recipient T cells. TCR-transduced T cells efficiently lysed primary B-cell leukemia, mantle cell lymphoma, and multiple myeloma in vitro. We also observed recognition and lysis of healthy BOB1-expressing B cells. In addition, strong BOB1-specific proliferation could be demonstrated for TCR-modified T cells upon antigen encounter. Furthermore, clear in vivo antitumor reactivity was observed of BOB1-specific TCR-engineered T cells in a xenograft mouse model of established multiple myeloma. Absence of reactivity toward a broad panel of BOB1− but HLA-B*07:02+ nonhematopoietic and hematopoietic cells indicated no off-target toxicity. Therefore, administration of BOB1-specific TCR-engineered T cells may provide novel cellular treatment options to patients with B-cell malignancies, including multiple myeloma.
Introduction
Immunotherapy for cancer based on the administration of chimeric antigen receptor (CAR)–modified T cells or monoclonal antibodies (mAbs) has demonstrated great clinical efficacy. In cases of hematological malignancies, CAR-engineered T cells specific for CD19- and CD20-targeting mAbs have led to complete remissions in patients with chronic lymphocytic leukemia (CLL), acute lymphoblastic leukemia (ALL), and follicular and diffuse large B-cell lymphoma.1-5 Therapeutic reactivity of mAbs and CAR-modified T cells is exerted by targeting extracellular cell-surface antigens. In contrast to mAbs and CARs, T-cell receptors (TCRs) recognize antigen-derived peptides that are bound to HLA molecules on the cell surface. Because HLA molecules constantly sample the entire endogenous proteome of cells, both extracellular and intracellular antigens are presented. Therefore, the potential pool of antigens targeted by T cells via their TCR is greater than the pool of target antigens accessible for mAbs or CARs.
Similar to CARs, T cells can be modified with a TCR of choice to equip T cells with a specific reactivity toward a well-defined antigen.6-9 Administration of such TCR-modified T cells is commonly referred to as TCR gene therapy and has already been successfully applied in the treatment of solid tumors.10,11 However, the widespread application of TCR gene therapy is hampered by lack of TCRs targeting common tumor-expressed self-antigens. Negative selection during thymic development deletes T cells carrying high-affinity TCRs for self-antigens in the context of autologous (self) HLA to prevent autoimmune reactions. In contrast, self-antigens presented in the context of allogeneic (nonself) HLA (alloHLA) molecules can elicit strong immune responses, as we have demonstrated by the isolation of PRAME-specific T cells after HLA-A*02:01–mismatched allogeneic stem-cell transplantation.12 Several strategies have been developed to generate T-cell responses toward specific self-antigens by exploiting the immunogenicity of alloHLA molecules.13-17
Despite the various technical advances in engineering T cells to recognize specific antigens, immunotherapy by administration of these modified T cells is lacking for many incurable malignancies, such as multiple myeloma (MM). Currently targeted antigens like CD20 and CD19 are not expressed by MM. In this study, we isolated from the allorepertoire of healthy individuals TCRs specific for the intracellular B cell–specific transcription factor BOB1 (also known as Oct coactivator from B cells [OCA-B] or Oct-binding factor-1 [OBF-1]). We demonstrate that TCR gene transfer of these BOB1-specific TCRs installs potent reactivity on recipient T cells, resulting in lysis of primary B-cell leukemia, mantle cell lymphoma (MCL), and MM in the absence of reactivity toward BOB1− cells. TCR-engineered T cells showed clear in vivo antitumor reactivity in a xenograft mouse model of established MM. Administration of BOB1-specific TCR-engineered T cells can bring novel immunotherapeutic treatments to patients with B-cell malignancies, including MM.
Materials and methods
Culture conditions and cells
Peripheral blood was obtained from different individuals after informed consent. Peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll-gradient centrifugation and were cryopreserved. T cells were cultured in T-cell medium consisting of IMDM (Lonza, Basel, Switzerland) supplemented with 100 IU/mL interleukin-2 (Proleukine; Novartis Pharma, Arnhem, The Netherlands), 5% fetal bovine serum (Gibco; Life Technologies, Carlsbad, CA), and 5% human serum. Isolation, generation, and culture of various hematopoietic and nonhematopoietic cell subsets and lines are described in the supplemental Material, available on the Blood Web site.
Isolation of BOB1-reactive T-cell clones
T cells binding to BOB1-specific peptide–major histocompatibility complex (pMHC) tetramers were isolated from PBMCs of healthy HLA-A2−/B7− individuals. PBMCs were first incubated with phycoerythrin (PE)-labeled pMHC tetramers for 1 hour at 4°C. Cells were washed twice and incubated with anti-PE microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) for 15 minutes at 4°C. PE-labeled cells were isolated on an LS Column (Miltenyi Biotec) according to manufacturer’s instruction. Positively selected cells were stained with an antibody against CD8 in combination with antibodies against CD4, CD14, and CD19. pMHC tetramer+ CD8+ T cells were single-cell sorted into round-bottom 96-well plates containing 5 × 104 irradiated (35 Gy) feeders in 100-µL T-cell medium supplemented with 0.8 µg/mL phytohemagglutinin.
TCR gene transfer to recipient CD4+ and CD8+ T cells
TCR variable α and TCR variable β usage of clone 4G11 was determined using reverse transcriptase polymerase chain reaction (PCR) and sequencing.18 A retroviral vector was constructed on an MP71 backbone with a codon-optimized and cysteine-modified TCRα and TCRβ chain joined by the T2A sequence in combination with the truncated nerve growth factor receptor (NGF-R) and ordered from GenScript (Piscataway, NJ). For in vivo experiments, the optimized TCRα and TCRβ chains joined by the T2A sequence were cloned and expressed on the SFG retroviral backbone.19
Purified CD4+ or CD8+ T cells were activated using irradiated autologous PBMCs and phytohemagglutinin. T cells were transduced using retroviral supernatant, and high-purity TCR-transduced T-cell populations were obtained as described in supplemental data.
FACS analysis
Fluorescence-activated cell sorting (FACS) was performed on an LSR II flow cytometer (BD Biosciences, Franklin Lakes, NJ) and analyzed using Diva (BD Biosciences) or FlowJo software (TreeStar, Ashland, OR). Isolated T-cell clones were analyzed for binding to specific pMHC tetramers and CD8 expression by staining with PE- or allophycocyanin-labeled pMHC tetramers and an Alexa700-conjugated antibody against CD8 (Invitrogen/Calteg, Buckingham, United Kingdom) combined with fluorescein isothiocyanate–labeled antibodies against CD4, CD14, and CD19 (BD Pharmingen, San Jose, CA). 20,000 cells of a T-cell clone were first incubated with 2-µg/mL pMHC tetramers for 15 minutes at 37°C before antibodies were added and incubated for an additional 15 minutes at 4°C. Similarly, 25,000 TCR-transduced or mock-transduced T cells were incubated with 2-µg/mL pMHC tetramers for 15 minutes at 37°C before antibodies against CD8, CD4, and CD271 (NGF-R; Sanbio, Uden, The Netherlands) were added and incubated at 4°C for 15 minutes. PBMCs, purified hematopoietic cell subsets, or activated cells were stained with antibodies against CD3, CD4, CD14, CD19, and CD34 (BD Pharmingen) at 4°C for 15 minutes.
Functional analysis and cytotoxicity assay
T cells and stimulator cells were coincubated for 18 hours before cell viability was assessed or supernatants were harvested, and interferon gamma (IFN-γ) or granulocyte-macrophage colony-stimulating factor production was measured by enzyme-linked immunosorbent assay (Sanquin Reagents or R&D Systems, Minneapolis, MN, respectively).
In vivo U266 MM model for antitumor efficacy
NSG mice (Jackson Laboratory, Bar Harbor, ME) were intravenously injected with 2 × 106 HLA-B7pos U266 cells expressing green fluorescent protein and luciferase. On day 21 after tumor inoculation, NSG mice were treated intravenously with 5 × 106 TCR 4G11 or mock-transduced T cells. Tumor growth was monitored using bioluminescence. On day 34, mice were euthanized, and bone marrow was harvested and analyzed for the presence of green fluorescent protein+ U266 cells by FACS analysis. Treatment groups were compared using unpaired 2-tailed Mann-Whitney test in GraphPad Prism (version 6.05; GraphPad Software, Inc., La Jolla, CA).
Results
Identification of BOB1 T-cell epitopes
We identified POU2AF1 as a candidate gene with B-cell lineage-restricted expression by comparing mRNA expression between CD19+ B cells and healthy hematopoietic and nonhematopoietic cell samples in an in-house–generated microarray gene expression database (supplemental Figure 1). POU2AF1 expression was high in primary CD19+ B cells, Epstein-Barr virus–transformed lymphoblastic B-cell lines (B-LCLs), and various B-cell malignancies including CLL, ALL, MCL, and MM. No expression of POU2AF1 was observed in CD34+ hematopoietic progenitor cells, T cells, professional antigen presenting cells, or other hematopoietic cell subsets not belonging to the B-cell compartment. Furthermore, expression was absent in nonhematopoietic cells, including fibroblasts, keratinocytes, melanocytes, and samples of the gastrointestinal tract. mRNA expression measured by microarray analysis was verified for a selected set of cell samples by real-time quantitative PCR (RT-qPCR; data not shown). Because POU2AF1 encodes for transcription factor BOB1, we mined the previously described B-LCL–derived HLA ligandome20 for naturally processed and major histocompatibility complex (MHC)-presented peptides of BOB1. Four peptides were identified, of which one was assigned to bind to HLA-A*02:01 (HLA-A2) and 3 peptides to HLA-B*07:02 (HLA-B7) using a public prediction algorithm21,22 (supplemental Table 1). Efficient binding to their respective HLA molecules and formation of stable pMHC monomers was validated using UV-exchange technology (supplemental Figure 2). Peptide BOB1245 efficiently bound to HLA-A2 and prevented MHC degradation comparable to human cytomegalovirus-derived peptide pp65NLV, which is known as a strong HLA-A2 ligand (supplemental Figure 2A). Similarly, peptides BOB1197, BOB144, and BOB114 prevented UV-induced MHC degradation of HLA-B7 comparable to peptide pp65TRP, which is a strong HLA-B7 ligand (supplemental Figure 2B). Stable pMHC tetramers for each peptide were generated by conjugating biotinylated pMHC monomers to fluorophore-labeled streptavidin.
Isolation of BOB1-reactive T-cell clones by massive pMHC tetramer enrichment
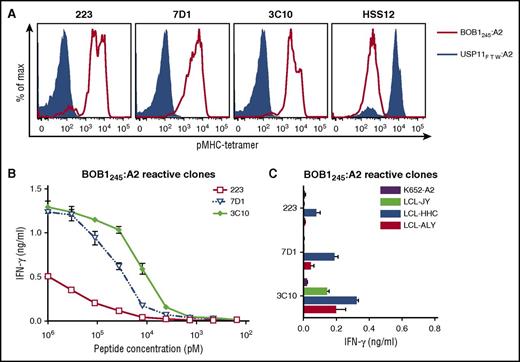
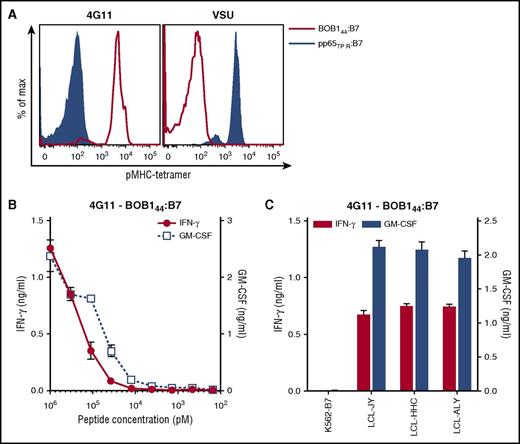
T-cell clones from HLA-A2−/B7− individuals were isolated using the generated pMHC tetramers composed of BOB1-derived peptides bound to HLA-A2 or HLA-B7. Between 250 and 1000 × 106 PBMCs from 6 healthy HLA-A2−/B7− individuals were labeled with the pool of pMHC tetramers. pMHC tetramer+ cells were first enriched by magnetic-activated cell sorting followed by single-cell sorting of pMHC tetramer+ CD8+ T cells. Thousands of T cells were clonally expanded for 2 weeks and screened in a high-throughput fashion for peptide specificity. To this aim, granulocyte-macrophage colony-stimulating factor secretion by the T-cell clones was assessed after stimulation with the BOB1− K562 cell line expressing either HLA-A*02:01 (K562-A2) or HLA-B*07:02 (K562-B7) in the absence or presence of exogenously loaded BOB1 peptides. A majority of T-cell clones lacked BOB1 specificity and recognized K562-A2 or K562-B7 cells independent of the presence of BOB1 peptides as exemplified by T-cell clones 3 and 29 (supplemental Figure 3). Other T-cells clones such as 227 showed no cytokine release under any stimulatory conditions, indicating insufficient sensitivity to recognize MHC-bound BOB1 peptides. In contrast, T-cell clones could be isolated demonstrating BOB1 specificity by recognizing either K562-A2 or K562-B7 cells when pulsed with BOB1 peptide. For example, T-cell clone 3C10 specifically recognized K562-A2 loaded with BOB1 peptide but did not react toward unloaded K562-A2 cells. Similarly, T-cell clone 4G11 reacted weakly but specifically toward K562-B7 cells pulsed with BOB1 peptides, whereas no reactivity against unloaded K562-B7 cells was observed. We performed pMHC tetramer staining to identify which of the four BOB1 peptides was recognized. No T-cell clone stained with pMHC tetramer BOB114:B7 or BOB1197:B7. However, 10 T-cell clones specifically bound to pMHC tetramer BOB1245:A2 (Figure 1A; supplemental Figure 4A), and one clone (4G11) specifically bound to pMHC tetramer BOB144:B7 (Figure 2A). The identified T-cell clones did not stain with an HLA-matched control pMHC tetramer containing an irrelevant peptide. Thus, from 6 HLA-A2−/B7− healthy individuals, 11 T-cell clones specific for BOB1 peptides presented either in HLA-A2 or HLA-B7 were selected for further analysis.
BOB1-reactive T-cell clones exhibit varying degrees of peptide sensitivity and avidity. BOB1-reactive T-cell clones recognizing peptide BOB1245 presented in HLA-A2 were assessed for peptide sensitivity by peptide titration and their capacity to recognize endogenously processed peptide. (A) Shown are histograms of 3 representative T-cell clones stained with pMHC tetramer BOB1245:A2 (red line) or control pMHC tetramer composed of irrelevant USP11-derived peptide FTWEGLYNV bound to HLA-A2 (USP11FTW:A2; blue area). T-cell clone HSS12 specific for USP11FTW:A2 served as control. (B-C) BOB1-reactive T-cell clones were coincubated with K562-A2 cells pulsed with (B) titrated BOB1245 peptide or (C) 3 BOB1-expressing HLA-A2+ B-LCLs. BOB1− K562-A2 cells were used as control. Shown are means with standard deviations of 1 representative experiment carried out in duplicate.
BOB1-reactive T-cell clones exhibit varying degrees of peptide sensitivity and avidity. BOB1-reactive T-cell clones recognizing peptide BOB1245 presented in HLA-A2 were assessed for peptide sensitivity by peptide titration and their capacity to recognize endogenously processed peptide. (A) Shown are histograms of 3 representative T-cell clones stained with pMHC tetramer BOB1245:A2 (red line) or control pMHC tetramer composed of irrelevant USP11-derived peptide FTWEGLYNV bound to HLA-A2 (USP11FTW:A2; blue area). T-cell clone HSS12 specific for USP11FTW:A2 served as control. (B-C) BOB1-reactive T-cell clones were coincubated with K562-A2 cells pulsed with (B) titrated BOB1245 peptide or (C) 3 BOB1-expressing HLA-A2+ B-LCLs. BOB1− K562-A2 cells were used as control. Shown are means with standard deviations of 1 representative experiment carried out in duplicate.
T-cell clone 4G11 recognizes endogenously processed and presented BOB144. (A) Shown are histograms of T-cell clone 4G11 or a control T-cell clone VSU specific for cytomegalovirus-derived peptide TPRVTGGGAM (pp65TPR). T-cell clones were stained with pMHC tetramer BOB144:B7 or pp65TPR:B7. (B-C) T-cell clone 4G11 was cocultured with K562-B7 pulsed with (B) titrated concentration of BOB144-peptide or (C) 3 BOB1-expressing HLA-B7+ B-LCLs. Shown are means with standard deviations of 1 experiment carried out in duplicate. GM-CSF, granulocyte-macrophage colony-stimulating factor.
T-cell clone 4G11 recognizes endogenously processed and presented BOB144. (A) Shown are histograms of T-cell clone 4G11 or a control T-cell clone VSU specific for cytomegalovirus-derived peptide TPRVTGGGAM (pp65TPR). T-cell clones were stained with pMHC tetramer BOB144:B7 or pp65TPR:B7. (B-C) T-cell clone 4G11 was cocultured with K562-B7 pulsed with (B) titrated concentration of BOB144-peptide or (C) 3 BOB1-expressing HLA-B7+ B-LCLs. Shown are means with standard deviations of 1 experiment carried out in duplicate. GM-CSF, granulocyte-macrophage colony-stimulating factor.
Selection of T-cell clones specific for BOB1245:A2 and BOB144:B7
Next, we aimed to identify high-avidity clones among a set of T-cell clones sharing the same peptide specificity. Sensitivity to BOB1 peptides was determined by coincubating T-cell clones with either K562-A2 or K562-B7 cells pulsed with titrated peptide. In addition, recognition of endogenously processed and presented peptide was assessed by coculturing T-cell clones with BOB1-expressing HLA-A2+/B7+ B-LCLs. Varying degrees of peptide sensitivity could be observed among T-cell clones classified as BOB1245:A2 reactive (Figure 1B). Representative T-cell clones 223, 7D1, and 3C10 demonstrated peptide specificity with increasing sensitivity. Increased sensitivity directly translated to better recognition of the 3 B-LCLs (Figure 1C). Clone 3C10 produced cytokine upon stimulation with all 3 B-LCLs, whereas less peptide-sensitive clones 223 and 7D1 showed no or only weak recognition of the 3 B-LCLs. Supplemental Figure 4B-C summarizes the analysis of other BOB1245-reactive T-cell clones. T-cell clone 4G11 exhibited BOB144 peptide–dependent recognition of K562-B7 cells (Figure 2B) and efficiently recognized all 3 B-LCLs (Figure 2C). These data indicated sufficient avidity of T-cell clone 4G11 to recognize endogenously processed and presented peptide.
In summary, from a pool of T-cell clones directed against different BOB1-derived peptides, 2 T-cell clones were selected by demonstrating specific recognition of peptide-pulsed target cells as well as reactivity against endogenously processed and presented peptide. T-cell clone 3C10 specifically recognized BOB1245 in HLA-A2, whereas T-cell clone 4G11 was specific for BOB144 presented in HLA-B7.
Specific recognition of malignant and healthy B-cell compartment
To further investigate the recognition profile of our selected BOB1-specific T-cell clones, we tested their reactivity toward primary BOB1-expressing B-cell malignancies. T-cell clone 4G11 efficiently recognized all HLA-B7+ primary B-cell malignancies tested, including 5 CLLs, 4 ALLs, 3 MCLs, and 4 MMs (Figure 3A). Furthermore, BOB1-dependent recognition of clone 4G11 could be demonstrated by introducing full-length BOB1 into K562-B7 cells (Figure 3C). T-cell clone 3C10 specific for BOB1245 presented in HLA-A2 also recognized primary BOB1-expressing B-cell malignancies, including MM. However, not all HLA-A2+ samples were recognized (Figure 3B). Clone 3C10 also failed to recognize K562-A2 when transduced with BOB1, indicating insufficient avidity to efficiently recognize endogenously processed peptide (Figure 3D). Two control T-cell clones recognizing household gene–derived peptides in the context of HLA-B*07:02 or HLA- A*02:01 confirmed sufficient and correct HLA subtype expression of all tested malignant cell samples (supplemental Figure 5A-B).
T-cell clone 4G11 efficiently recognizes B-cell malignancies while sparing non–B-cell lineages. (A-B) T-cell clones (A) 4G11 and (B) 3C10 were coincubated with primary samples of malignant B cells, including CLL, ALL, MCL, and MM. HLA allotype of each sample is indicated. Controls included BOB1− K562-B7 cells and BOB1-expressing LCL-JY. Shown is 1 representative experiment of 2 independent experiments. (C-D) T-cell clones (C) 4G11 and (D) 3C10 were incubated with K562-B7 and K562-A2 cells alone or transduced to express BOB1 (K562-B7 + BOB1 or K562-A2 + BOB1, respectively) or K562-B7 and K562-A2 cells pulsed with 1 µM BOB144 or BOB1245 peptide, respectively. (E-J) T-cell clone 4G11 was cocultured with different HLA-B7+ healthy primary or activated cell subsets. (E) Fibroblasts (FBs) from 3 different individuals were either left untreated or cultured for 4 days in 200 IU/ml IFN-γ (FB + IFN-γ) before coculture. Primary bronchial epithelial cells were cultured under (F) non–air-exposed or (G) air-exposed condition before being used as stimulator cells. Cells had also been cultured in the absence (−IFN-γ) or presence (+IFN-γ) of 100 IU/ml IFN-γ for 2 days before coculture. (H) Clone 4G11 was coincubated with various HLA-B7+ BOB1− nonhematopoietic tumor cell lines. (I) Primary CD19+, CD4+, CD14+, and CD34+ cells were isolated from PBMCs of healthy individuals (UDR, UHK, AFD, and ALY) and cocultured in different responder-to-stimulator (R:S) ratios. Activated B cells (CD19 [CD40L]) were generated by stimulating CD19+ B cells with CD40L. Activated T cells were generated from phytohemagglutinin-stimulated PBMCs. (J) Immature (imDC) and mature (mDC) dendritic cells were monocyte derived. (K) T-cell clone 4G11 was cocultured with a panel of BOB1-expressing B-LCLs expressing a wide variety of HLA class I and II molecules. HLA-B7 status of each B-LCL is indicated with + (positive) or – (negative). IFN-γ production was measured after 18 hours of coculture. Shown are means with standard deviations of 1 experiment carried out in duplicate. Representative data from ≥2 independent experiments.
T-cell clone 4G11 efficiently recognizes B-cell malignancies while sparing non–B-cell lineages. (A-B) T-cell clones (A) 4G11 and (B) 3C10 were coincubated with primary samples of malignant B cells, including CLL, ALL, MCL, and MM. HLA allotype of each sample is indicated. Controls included BOB1− K562-B7 cells and BOB1-expressing LCL-JY. Shown is 1 representative experiment of 2 independent experiments. (C-D) T-cell clones (C) 4G11 and (D) 3C10 were incubated with K562-B7 and K562-A2 cells alone or transduced to express BOB1 (K562-B7 + BOB1 or K562-A2 + BOB1, respectively) or K562-B7 and K562-A2 cells pulsed with 1 µM BOB144 or BOB1245 peptide, respectively. (E-J) T-cell clone 4G11 was cocultured with different HLA-B7+ healthy primary or activated cell subsets. (E) Fibroblasts (FBs) from 3 different individuals were either left untreated or cultured for 4 days in 200 IU/ml IFN-γ (FB + IFN-γ) before coculture. Primary bronchial epithelial cells were cultured under (F) non–air-exposed or (G) air-exposed condition before being used as stimulator cells. Cells had also been cultured in the absence (−IFN-γ) or presence (+IFN-γ) of 100 IU/ml IFN-γ for 2 days before coculture. (H) Clone 4G11 was coincubated with various HLA-B7+ BOB1− nonhematopoietic tumor cell lines. (I) Primary CD19+, CD4+, CD14+, and CD34+ cells were isolated from PBMCs of healthy individuals (UDR, UHK, AFD, and ALY) and cocultured in different responder-to-stimulator (R:S) ratios. Activated B cells (CD19 [CD40L]) were generated by stimulating CD19+ B cells with CD40L. Activated T cells were generated from phytohemagglutinin-stimulated PBMCs. (J) Immature (imDC) and mature (mDC) dendritic cells were monocyte derived. (K) T-cell clone 4G11 was cocultured with a panel of BOB1-expressing B-LCLs expressing a wide variety of HLA class I and II molecules. HLA-B7 status of each B-LCL is indicated with + (positive) or – (negative). IFN-γ production was measured after 18 hours of coculture. Shown are means with standard deviations of 1 experiment carried out in duplicate. Representative data from ≥2 independent experiments.
Because T-cell clone 4G11 efficiently recognized all different B-cell malignancies tested, we further investigated the recognition profile of this clone by coincubation with various HLA-B7+ nonhematopoietic and hematopoietic cell subsets. T-cell clone 4G11 did not react with BOB1− fibroblasts even when cultured with IFN-γ to mimic inflammation (Figure 3E). Additionally, we tested clone 4G11 for the recognition of primary bronchial epithelial cells (PBECs), because a recent publication reported on the expression of POU2AF1 in human airway epithelium.23 POU2AF1 mRNA could be detected at very low levels in air-exposed and non–air-exposed PBECs by RT-qPCR (data not shown). However, clone 4G11 did not produce IFN-γ upon stimulation with these cells (Figure 3F-G). Recognition was also absent when PBECs had been incubated with IFN-γ to mimic inflammation (Figure 3F-G). Clone 4G11 did not produce cytokine when stimulated with a panel of different BOB1− nonhematopoietic tumor cell lines (Figure 3H). Absence of BOB1 expression in tumor cell lines was confirmed by RT-qPCR (data not shown). Furthermore, reactivity toward hematopoietic cell subsets was restricted to the B-cell compartment. BOB1-expressing primary and activated B cells were recognized, whereas no reactivity was found against BOB1− resting and activated T cells, monocytes, or monocyte-derived immature or mature dendritic cells derived from 2 different HLA-B7+ donors or CD34+ hematopoietic progenitor cells (Figure 3I-J). A control clone CTZ, specific for a ubiquitously expressed antigen in the context of HLA-B7, efficiently recognized all cell samples tested, confirming the stimulatory capacity of these cells (supplemental Figure 5C-H).
Finally, when tested against a panel of BOB1-expressing B-LCLs expressing 95% of common and rare HLA class I and II alleles,24 T-cell clone 4G11 only reacted to HLA-B7+ B-LCLs, indicating HLA-B7 restriction without cross-reactivity to all other HLA class I alleles tested (Figure 3K; supplemental Table 2).
In conclusion, T-cell clone 4G11 showed potent antitumor reactivity as demonstrated by the recognition of malignant B cells, including MM. Furthermore, absence of reactivity toward healthy hematopoietic non–B cells and nonhematopoietic cells indicated no off-target toxicity.
TCR gene transfer installs BOB1 reactivity onto recipient T cells
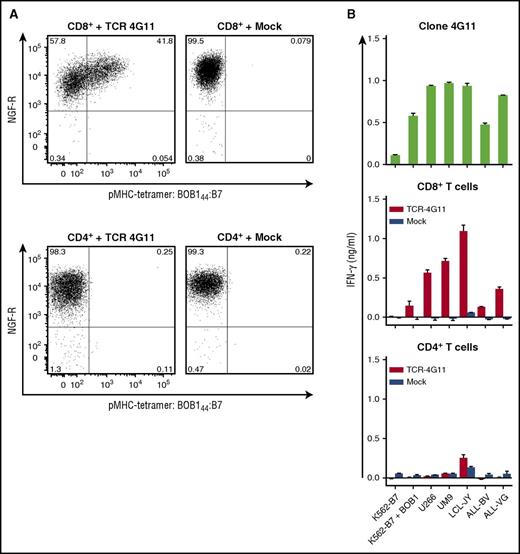
Next, we investigated whether gene transfer of the TCR of clone 4G11 (TCR-4G11) could install BOB1 reactivity onto recipient cells. TCR-4G11 was sequenced, codon optimized, modified with a disulfide bond to increase preferential pairing of the TCRα and TCRβ chain, and cloned into the MP71 vector expressing NGF-R as a marker gene. Expression of NGF-R was used to enrich TCR-transduced T cells to high purity (>98%) by magnetic-activated cell sorting–guided isolation of NGF-R–expressing cells. Retrovirally transduced CD8+ T cells from an HLA-B7+ healthy individual expressed TCR-4G11 on the cell surface, indicated by their capacity to bind pMHC tetramer BOB144:B7 (Figure 4A). More intensive staining with pMHC tetramer correlated with higher NGF-R expression, suggesting that cells expressing higher levels of the introduced TCR bound pMHC tetramer more efficiently. Tetramer binding was not observed for mock-transduced T cells. Although CD4+ T cells could be transduced, as indicated by the expression of NGF-R, no binding to BOB144:B7 tetramer was observed (Figure 4A). TCR-transduced CD8+ T cells readily recognized BOB1-expressing HLA-B7+ stimulator cells, such as MM cell lines UM9 and U266, LCL-JY, and two ALL cell lines (ALL-BV and ALL-VG), mirroring the reactivity profile of T-cell clone 4G11 (Figure 4B). In contrast, TCR-transduced CD4+ T cells failed to recognize any of these cell lines. No recognition for mock-transduced CD8+ T cells was observed, indicating that recognition was mediated by the introduced TCR-4G11.
Transfer of TCR-4G11 installs BOB1 reactivity on recipient CD8+ T cells. CD4+ and CD8+ T cells were isolated from a healthy HLA-B7+ individual using magnetic-activated cell sorting (MACS). T cells were transduced with retroviral supernatant to express TCR-4G11 together with NGF-R. Transduction with an empty vector (mock) containing only the NGF-R marker gene served as control. Transduced T cells were purified based on the expression of marker gene NGF-R using MACS. Level of purity exceeded 98% in all cases. (A) Shown are FACS plots of purified T cells after transduction. CD8+ (top row) or CD4+ (bottom row) T cells were stained with pMHC tetramer BOB144:B7 and an antibody against NGF-R. Numbers in corners indicate percentage of cells per quadrant. FACS plots are shown with biexponential axes. (B) T-cell clone 4G11 or purified transduced CD8+ or CD4+ T cells were coincubated with various HLA-B7+ cell lines. Cell lines included K562-B7 transduced to express BOB1 (K562-B7 + BOB1), 2 MM cell lines UM9 and U266, LCL-JY, and 2 ALL cell lines ALL-BV and ALL-VG. IFN-γ concentration was assessed after 18 hours of coculture. Shown are means with standard deviations of 1 experiment carried out in duplicate.
Transfer of TCR-4G11 installs BOB1 reactivity on recipient CD8+ T cells. CD4+ and CD8+ T cells were isolated from a healthy HLA-B7+ individual using magnetic-activated cell sorting (MACS). T cells were transduced with retroviral supernatant to express TCR-4G11 together with NGF-R. Transduction with an empty vector (mock) containing only the NGF-R marker gene served as control. Transduced T cells were purified based on the expression of marker gene NGF-R using MACS. Level of purity exceeded 98% in all cases. (A) Shown are FACS plots of purified T cells after transduction. CD8+ (top row) or CD4+ (bottom row) T cells were stained with pMHC tetramer BOB144:B7 and an antibody against NGF-R. Numbers in corners indicate percentage of cells per quadrant. FACS plots are shown with biexponential axes. (B) T-cell clone 4G11 or purified transduced CD8+ or CD4+ T cells were coincubated with various HLA-B7+ cell lines. Cell lines included K562-B7 transduced to express BOB1 (K562-B7 + BOB1), 2 MM cell lines UM9 and U266, LCL-JY, and 2 ALL cell lines ALL-BV and ALL-VG. IFN-γ concentration was assessed after 18 hours of coculture. Shown are means with standard deviations of 1 experiment carried out in duplicate.
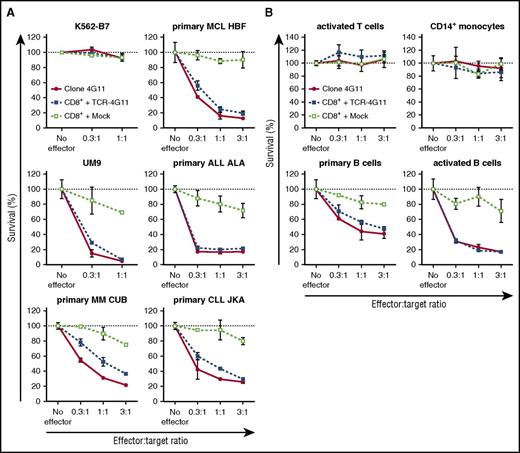
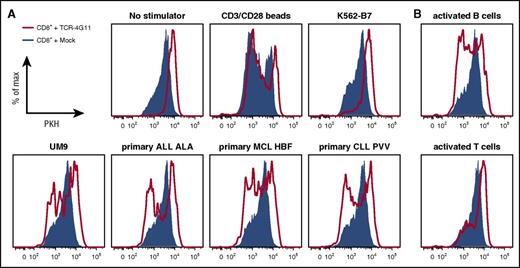
Furthermore, TCR-transduced CD8+ T cells efficiently lysed primary HLA-B7+ malignant cell samples (Figure 5A; supplemental Figure 6A). Complete or nearly complete lysis was observed for primary ALL and MCL samples and MM cell lines at equal effector-to-target ratios and even when targets cells exceeded effector cells 3-fold. Efficient lysis by TCR-transduced T cells could also be observed for both tested primary CLL samples as well as both ALL cell lines. In addition, both purified HLA-B7+ primary MM samples were readily lysed by TCR-transduced T cells at low effector-to-target ratios. In all cases, no lysis was observed for mock-transduced T cells, indicating that lysis was mediated by the introduced TCR-4G11. BOB1 expression in healthy B cells also led to lysis by autologous TCR-transduced CD8+ T cells (Figure 5B). Lysis was specific, because the BOB1− K562-B7 cell line was not lysed (Figure 5A). In addition, TCR-transduced CD8+ T cells did not lyse autologous activated T cells or CD14+ monocytes, indicating a safe reactivity profile (Figure 5B). Finally, TCR-transduced CD8+ T cells proliferated upon stimulation with various BOB1-expressing HLA-B7+ primary samples, including ALL, CLL, MCL, and MM cell lines and autologous activated B cells (Figure 6A-B; supplemental Figure 6B). In contrast, absence of any stimulus or stimulation with antigen-negative cell line K562-B7 or autologous activated T cells did not lead to proliferation of TCR-transduced T cells. Proliferative capacity of TCR- and mock-transduced T cells was confirmed using CD3/CD28 T-cell activator beads.
TCR-transduced CD8+ T cells efficiently lyse primary B-cell malignancies, including MM. (A-B) T-cell clone 4G11 or purified TCR- or mock-transduced CD8+ T cells were tested for their lytic capacity of HLA-B7+ target cells. PKH-labeled target cells were cocultured at various effector-to-target ratios with effector T cells. After 18 hours of coculture, the number of live targets cells was assessed by flow cytometry, and percent survival was calculated. (A) Malignant cell samples included MM cell line UM9, primary MM, MCL, ALL, and CLL. Controls included BOB1− cell line K562-B7. (B) Healthy hematopoietic cells were of the same origin as transduced T cells (autologous setting) and included phytohemagglutinin-activated T cells, CD14+ monocytes, CD19+ primary B cells, and CD40L-activated B cells. Shown are means with standard deviations of 1 experiment carried out in triplicate.
TCR-transduced CD8+ T cells efficiently lyse primary B-cell malignancies, including MM. (A-B) T-cell clone 4G11 or purified TCR- or mock-transduced CD8+ T cells were tested for their lytic capacity of HLA-B7+ target cells. PKH-labeled target cells were cocultured at various effector-to-target ratios with effector T cells. After 18 hours of coculture, the number of live targets cells was assessed by flow cytometry, and percent survival was calculated. (A) Malignant cell samples included MM cell line UM9, primary MM, MCL, ALL, and CLL. Controls included BOB1− cell line K562-B7. (B) Healthy hematopoietic cells were of the same origin as transduced T cells (autologous setting) and included phytohemagglutinin-activated T cells, CD14+ monocytes, CD19+ primary B cells, and CD40L-activated B cells. Shown are means with standard deviations of 1 experiment carried out in triplicate.
TCR-transduced CD8+ T cells proliferate upon antigen encounter. (A-B) PKH-labeled transduced CD8+ T cells were cocultured with irradiated HLA-B7+ stimulator cells. Histograms show TCR-transduced (red line) or mock-transduced (blue area) CD8+ T cells after 5 days of coculture. (A) Stimulator cells included cell line UM9 and primary ALL, CLL, and MCL. Negative controls included culture in absence of stimulator cells (no stimulators) or coculture with BOB1− K562-B7 cells. Positive control included stimulation in the presence of CD3/CD28 T-cell activator beads (CD3/CD28 beads). (B) Autologous CD40L-stimulated B cells or phytohemagglutinin-activated T cells were used as stimulator cells.
TCR-transduced CD8+ T cells proliferate upon antigen encounter. (A-B) PKH-labeled transduced CD8+ T cells were cocultured with irradiated HLA-B7+ stimulator cells. Histograms show TCR-transduced (red line) or mock-transduced (blue area) CD8+ T cells after 5 days of coculture. (A) Stimulator cells included cell line UM9 and primary ALL, CLL, and MCL. Negative controls included culture in absence of stimulator cells (no stimulators) or coculture with BOB1− K562-B7 cells. Positive control included stimulation in the presence of CD3/CD28 T-cell activator beads (CD3/CD28 beads). (B) Autologous CD40L-stimulated B cells or phytohemagglutinin-activated T cells were used as stimulator cells.
In summary, TCR gene transfer of TCR-4G11 installed BOB1 reactivity onto recipient CD8+ T cells. TCR-transduced T cells efficiently lysed primary ALL, CLL, MCL, and MM at low effector-to-target ratios while sparing non–B cells. Furthermore, TCR-transduced T cells readily proliferated upon antigen encounter.
In vivo efficacy of BOB1-specific TCR-transduced T cells
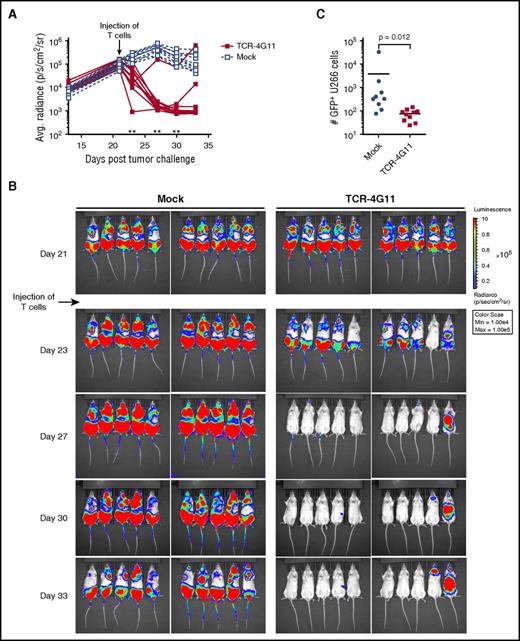
Using an MM xenograft mouse model, we investigated the in vivo efficacy of our TCR-transduced T cells. TCR-4G11–expressing T cells demonstrated strong antitumor reactivity against U266 cells that were allowed to expand in NSG mice for 3 weeks before treatment. Mock-transduced T cells failed to control tumor growth, although in later stages, tumor expansion was curbed, probably caused by the alloreactivity of mock-transduced T cells. In contrast, significant tumor reduction was observed in mice after injection of TCR-4G11 T cells (Figure 7A-B). When bone marrows of mice were investigated for the presence of tumor cells at the end of the experiment, mock-treated mice contained on average an order of magnitude more tumor cells than could be found in TCR-treated mice (Figure 7C).
TCR-transduced CD8+ T cells demonstrate in vivo antitumor efficacy in model of established MM. Green fluorescent protein (GFP)+ U266 MM cells expressing luciferase were allowed to engraft in NSG mice. Mice were intravenously treated with TCR-transduced (TCR-4G11) or mock-transduced (mock) T cells 21 days after tumor injection. (A-B) Tumor growth was monitored by bioluminescence. Each treatment group contained 10 mice. Results from unpaired 2-tailed Mann-Whitney test comparing differences in mean bioluminescence between treatment groups is shown in (A), **P < .01. (C) Mice were euthanized on day 33; bone marrow was harvested and analyzed for presence of GFP+ U266 cells. Result from unpaired 2-tailed Mann-Whitney test is shown.
TCR-transduced CD8+ T cells demonstrate in vivo antitumor efficacy in model of established MM. Green fluorescent protein (GFP)+ U266 MM cells expressing luciferase were allowed to engraft in NSG mice. Mice were intravenously treated with TCR-transduced (TCR-4G11) or mock-transduced (mock) T cells 21 days after tumor injection. (A-B) Tumor growth was monitored by bioluminescence. Each treatment group contained 10 mice. Results from unpaired 2-tailed Mann-Whitney test comparing differences in mean bioluminescence between treatment groups is shown in (A), **P < .01. (C) Mice were euthanized on day 33; bone marrow was harvested and analyzed for presence of GFP+ U266 cells. Result from unpaired 2-tailed Mann-Whitney test is shown.
In conclusion, BOB1-specific TCR-transduced T cells showed potent in vivo antitumor reactivity in an in vivo xenograft model of established MM.
Discussion
TCR gene transfer is an attractive strategy for cancer therapy that equips T cells with a TCR of defined antigen specificity to eradicate antigen-positive malignant cells. However, TCR gene therapy is still unavailable for many solid tumors and hematological malignancies, because no suitable targets or antigen-specific TCRs have been identified. Here, we describe the isolation of TCRs recognizing peptides derived from the intracellular transcription factor BOB1 presented in the context of HLA-A2 or HLA-B7.
BOB1 is highly expressed in B-cell malignancies, such as ALL, CLL, and MCL. Moreover, BOB1 is also highly expressed in MM, for which no curative treatment is currently available. BOB1 is an octamer-binding B cell–specific coactivator. By binding to the POUH and POUS subdomains of OCT-1/2 proteins, it increases their affinity for octamer-containing DNA and further increases octamer-dependent gene transcription.25,26 In addition, cytosolic BOB1 has been implicated to regulate early events in intracellular signaling of B cells via its interaction with tyrosine kinase SYK.27 A drastically reduced B-cell count resulting from increased apoptosis of B cells in the bone marrow and complete lack of formation of germinal centers in BOB1-knockout mice has established a broad role for BOB1 in B-cell development and function throughout all B-cell stages.28-32 The POU2AF1 gene locus has been identified as a target of gene amplification in a subset of MM.33 Furthermore, in MM cell lines showing gene amplification of the POU2AF1 gene locus, it has been shown that proliferation can be hampered by BOB1 knockdown or knockdown of downstream targets of BOB1. Based on the role of BOB1 in B-cell development and its involvement in proliferation in a subset of MM, it is tempting to speculate that BOB1 is a vital player in B-cell survival and that therefore BOB1-loss tumor escape variants may not likely occur in the course of BOB1-targeting therapy, as has been observed when targeting other antigens.2,34 However, whether B-cell malignancies indeed lack the capacity to escape from immune surveillance by shutting down BOB1 expression should become clear in clinical trials in which BOB1 is specifically targeted.
By exploiting the immunogenicity of alloHLA, we were able to identify a high-affinity TCR targeting the self-antigen BOB1. Although several T-cell clones directed against different BOB1-derived peptides were isolated, only T-cell clone 4G11 demonstrated sufficient peptide sensitivity as well as stringent antigen specificity. Endogenously processed peptide was efficiently and reproducibly recognized on HLA-B7+ B-LCLs as well as B-cell malignancies. In contrast, T-cell clone 3C10 specific for BOB1245 presented in HLA-A2 demonstrated similar peptide sensitivity but failed to recognize all HLA-A2+ B-LCLs and B-cell malignancies. These data may suggest superior affinity of TCR-4G11 for BOB144:B7 than TCR-3C10 for BOB1245:A2. However, the different recognition profiles of the T-cell clones may also stem from more efficient processing and greater abundance of BOB144:B7 on the cell surface as compared with BOB1245:A2. If true, efficient targeting of BOB1245 bound to HLA-A2 requires expression of a higher-affinity TCR than expressed by clone 3C10.
The various reactivity patterns of the different T-cell clones resemble our previous experience when using pMHC tetramers for enrichment of specific T-cell populations.17,35,36 Reactivity against antigen-negative K562 cell line was often observed and indicated potential harmful off-target toxicity. Of note, T-cell clone 17A5, demonstrating recognition of an irrelevant peptide in HLA-A2 when stimulated with BOB1− K562-A2, also recognized HLA-A2+ fibroblasts and healthy hematopoietic cell subsets (supplemental Figure 7).
T-cell clone 4G11 specific for BOB144 presented in the context of HLA-B7 strongly recognized various B-cell malignancies, including MM. Furthermore, T cells engineered to express TCR-4G11 successfully controlled tumor growth in an in vivo model of established MM. No reactivity against a wide panel of BOB1− but HLA-B7+ stimulator cells and no cross-reactivity with other HLA class I and II alleles were observed for TCR-4G11. From a standard protein BLAST search, no protein could be identified that shared 100% amino acid (aa) sequence similarity with peptide BOB144. Proteins with partial aa sequence similarity did not contain peptides covering this partial homology that would strongly interact with HLA-B7 as predicted by a public prediction algorithm.21,22 Additionally, proteins sharing a partial aa sequence with the BOB144 epitope are expressed in T cells or monocytes, which we have demonstrated not to be recognized by clone 4G11. However, the possibility remains that unwanted off-target or on-target off-tumor toxicity may occur once TCR-modified T cells are administered in patients.37,38 Therefore, TCR-engineered T cells should additionally be equipped with a suicide switch to abolish their reactivity in case of adverse effects.39-44 Furthermore, because healthy B cells expressing BOB1 will also be depleted in the course of therapy, specific depletion of BOB1-reactive TCR-modified T cells after successful therapy may be desirable to enable restoration of the healthy B-cell compartment. Nonetheless, long-term B-cell aplasia is tolerable from a clinical perspective and can be managed, as has been demonstrated by other immunotherapeutic interventions targeting B cell–expressed antigens.3,45
Transfer of TCR-4G11 installed BOB1 reactivity onto recipient CD8+ but not CD4+ T cells. We speculate that TCR-4G11 requires expression of coreceptor CD8 for sufficient binding to cognate pMHC molecules, as has been observed for other high-affinity TCRs.46,47 However, BOB1-specific CD4 T-cell help could be provided by co-transduction of TCR-4G11 and coreceptor CD8.47,48
In conclusion, we expect BOB1-targeting TCRs to be a valuable addition to current immunotherapies by broadening their application to yet incurable diseases like MM. Furthermore, because of broad expression of BOB1 throughout the entire B-cell lineage, BOB1-specific T cells could be of use in patients with B-cell malignancies in which standard therapy or other immunotherapies have failed. Moreover, targeting monomorphic (nonmutated) peptides in the context of common HLA molecules, BOB1-targeting TCRs are widely applicable by bringing novel therapeutic strategies to patients with tumors in which the occurrence of an endogenous immune response directed against mutated neoantigens is less likely.49
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Guido de Roo and Sabrina A. J. Veld (Department of Hematology, Leiden University Medical Center, Leiden, The Netherlands) for providing expert technical assistance in flow cytometric cell sorting. The UM9 cell line was a kind gift of Anniek van der Waart and Harry Dolstra (Department of Laboratory Medicine, Laboratory of Hematology, Radboud University Medical Center, Nijmegen, The Netherlands). The U266 cell line was kindly provided by Tuna Mutis (Department of Hematology, Free University Medical Center, Amsterdam, The Netherlands). The authors thank Aaron Foster and David Spencer (Bellicum Pharmaceuticals, Inc., Houston, TX) for valuable discussions and careful review of the manuscript.
This research was supported by the financial assistance of the Dutch Cancer Society (2010-4832) and the Landsteiner Foundation for Blood Transfusion Research (LSBR0713).
Authorship
Contribution: L.J. designed, performed, analyzed, and interpreted all experiments and wrote the manuscript; P.H. designed the study; R.S.H. determined TRAV and TRBV usage and constructed retroviral expression vectors and performed RT-qPCR; M.G.D.K. analyzed peptide elution studies and identified peptides; D.M.v.d.S. generated pMHC monomers and performed and analyzed experiments; T.R and T.P.-H. designed, performed, analyzed, and interpreted in vivo antitumor efficacy experiments; A.H.d.R. performed and analyzed mass spectrometry experiments; M.P.S. and M.H.M. performed experiments; M.G. provided the in-house mRNA expression database and helped in analysis of the database; P.A.v.V. designed and cosupervised the study; J.H.F.F. designed and supervised the study and wrote the manuscript; M.H.M.H. analyzed and interpreted experiments, designed and supervised the study, and wrote the manuscript. All authors revised and edited the manuscript.
Conflict-of-interest disclosure: T.R. and T.P.-H. are employees of Bellicum Pharmaceuticals, Inc. The remaining authors declare no competing financial interests.
Correspondence: Lorenz Jahn, Department of Hematology, Leiden University Medical Center, Postzone C2R-140, P.O. Box 9600, 2300 RC Leiden, The Netherlands; e-mail: l.jahn@lumc.nl; and Mirjam H. M. Heemskerk, Department of Hematology, Leiden University Medical Center, Postzone C2R-140, P.O. Box 9600, 2300 RC Leiden, The Netherlands; e-mail: m.h.m.heemskerk@lumc.nl.

![Figure 3. T-cell clone 4G11 efficiently recognizes B-cell malignancies while sparing non–B-cell lineages. (A-B) T-cell clones (A) 4G11 and (B) 3C10 were coincubated with primary samples of malignant B cells, including CLL, ALL, MCL, and MM. HLA allotype of each sample is indicated. Controls included BOB1− K562-B7 cells and BOB1-expressing LCL-JY. Shown is 1 representative experiment of 2 independent experiments. (C-D) T-cell clones (C) 4G11 and (D) 3C10 were incubated with K562-B7 and K562-A2 cells alone or transduced to express BOB1 (K562-B7 + BOB1 or K562-A2 + BOB1, respectively) or K562-B7 and K562-A2 cells pulsed with 1 µM BOB144 or BOB1245 peptide, respectively. (E-J) T-cell clone 4G11 was cocultured with different HLA-B7+ healthy primary or activated cell subsets. (E) Fibroblasts (FBs) from 3 different individuals were either left untreated or cultured for 4 days in 200 IU/ml IFN-γ (FB + IFN-γ) before coculture. Primary bronchial epithelial cells were cultured under (F) non–air-exposed or (G) air-exposed condition before being used as stimulator cells. Cells had also been cultured in the absence (−IFN-γ) or presence (+IFN-γ) of 100 IU/ml IFN-γ for 2 days before coculture. (H) Clone 4G11 was coincubated with various HLA-B7+ BOB1− nonhematopoietic tumor cell lines. (I) Primary CD19+, CD4+, CD14+, and CD34+ cells were isolated from PBMCs of healthy individuals (UDR, UHK, AFD, and ALY) and cocultured in different responder-to-stimulator (R:S) ratios. Activated B cells (CD19 [CD40L]) were generated by stimulating CD19+ B cells with CD40L. Activated T cells were generated from phytohemagglutinin-stimulated PBMCs. (J) Immature (imDC) and mature (mDC) dendritic cells were monocyte derived. (K) T-cell clone 4G11 was cocultured with a panel of BOB1-expressing B-LCLs expressing a wide variety of HLA class I and II molecules. HLA-B7 status of each B-LCL is indicated with + (positive) or – (negative). IFN-γ production was measured after 18 hours of coculture. Shown are means with standard deviations of 1 experiment carried out in duplicate. Representative data from ≥2 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/10/10.1182_blood-2016-09-737536/4/m_blood737536f3.jpeg?Expires=1768771710&Signature=xlqCBGxOHPKC0Uu6ok1s5xYU4O0IL8rBl7PnHmBLMbXQuZjFKQmoeFQtpYhRd1DEOnnaIG4IyEhtPYLc~8rXG417OC8duyODB8EBRz2eNRqPmXFHEl2OqRrQOJSGQmlRNBAh~VSDVrbESB7wCsQZ2ge5r1BVaMcRGE1~xhciIdf4kNbOSjCy4isfTTtZh0QP5qbTLgi~UcKaqQ2cLX96mIipbqaCYSqfzXJCejPH9PbAP4LDholwEr~ShZOjZfTuoyjINY1onomfsejryiNntSD0jTL0c5n8K2cPWTTqDwoT5rRVlqyh3FjoLbyHLX0xEM6ZfNAshHav8fVifm75GQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)