To the editor:
The diagnosis of chronic myelomonocytic leukemia (CMML), as defined by the World Health Organization (WHO), requires the persistent presence of peripheral monocytosis (≥1 × 109/L) accounting for ≥10% of the total white blood cell count (WBC).1 However, peripheral monocytosis is not pathognomonic of CMML and can be observed in other hematologic neoplasms and benign reactive conditions. Further, monocytes are now recognized as 3 functionally and immunophenotypically distinct cell populations. These populations include classical monocytes (immunophenotypically defined as CD14+/CD16−), intermediate monocytes (defined as CD14+/CD16+), and nonclassical monocytes (defined as CD14−/CD16+). Upon stimulation with lipopolysaccharide, classical monocytes produce the highest levels and broadest range of cytokines that include granulocyte colony-stimulating factor, interleukin 10 (IL-10), CCL2, and IL-6.2 Stimulated nonclassical subsets produce comparatively higher levels of tumor necrosis factor-α and IL-1B, and intermediate monocytes produce all of these cytokines at significantly lower levels.2
In humans, ∼90% of monocytes are comprised of a classical subset and 10% are comprised of nonclassical or intermediate monocytes.3 Recently, Selimoglu-Buet et al annotated the composition of monocyte subsets in patients with CMML. Utilizing flow cytometry to segregate monocyte subsets, the authors demonstrated that classical monocytes were uniformly increased in CMML.4 They further demonstrated that classical monocytosis (CM) was a highly specific marker capable of accurately distinguishing CMML from reactive conditions in a mixed cohort of hematologic neoplasms even without considering total monocyte count, genetic, or cytogenetic data.4 Although these data suggests that monocyte partitioning may serve as a compelling diagnostic marker for this disease, the aforementioned study has been the only report to date describing this observation. Further, given the close genetic and clinical association between myelodysplastic syndrome (MDS) and CMML, the diagnostic and clinical implication of monocyte subset in MDS is of importance and remains largely unexplored.
Here, we aimed to validate the findings published by Selimoglu-Buet et al in a clinical and genetically annotated cohort of normal age-matched controls, CMML patients, and specifically focus on the consequence of monocyte subsets in MDS. To achieve this, we generated a Clinical Laboratory Improvement Amendments (CLIA)-certified clinical monocyte subset test and prospectively profiled monocyte subsets in clinically and genetically annotated patients with a suspected diagnosis of CMML or MDS. Somatic mutation analysis was performed via a CLIA-certified inhouse 52-gene myeloid panel that evaluates all recurrently mutated genes in CMML (TruSight myeloid sequencing panel; Illumina, San Diego, CA). Descriptive statistics were used to summarize clinical demographics, genotyping, and their association to CM. Receiver operating curves (ROCs) were generated to test the sensitivity and specificity of the monocyte analysis and all P values calculated were two-sided. Furthermore, CM was defined as the presence of CD14+/CD16− monocytes comprising a total of at least 94% of total monocytes based on a cut point derived from analysis of 33 normal controls during CLIA certification.
In total, monocyte subsets were profiled in 159 cases with availability of somatic mutation analysis between October 2015 and May 2016. The pathologic diagnosis of patients in our cohort included CMML (n = 29), MDS (n = 86), other myeloid malignancies (n = 26), and reactive conditions (n = 18). The median relative CM in the CMML cohort was significantly greater at 97.85% (range, 83.01% to 99.85%) compared with the MDS cohort at 89.16% (range, 48.01% to 99.68%), and reactive conditions at 90.06% (range, 69.67% to 98.99%).
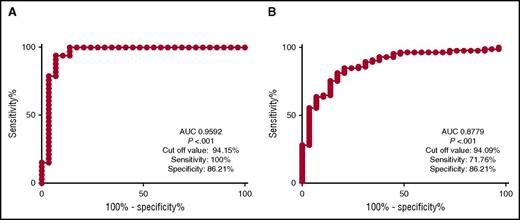
As previously reported, CM was evident in all CMML cases and was capable of differentiating CMML from normal age-matched controls with reactive monocytosis. ROC analysis confirmed that the assay was capable of differentiating these groups with a sensitivity of 100% and specificity of 86.21% (area under the curve [AUC] of 0.9592; P < .001) (Figure 1A). To determine if there were clinical and genetic CMML subgroups that can be stratified by monocyte subsets, we analyzed the pattern of classical, nonclassical, and intermediate monocytosis in all cases. However, no difference in CM was evident when stratifying cases by French-American-British (P = .09) or WHO-defined CMML subtypes (P = .56).
ROC analysis of CMML in comparison with reactive monocytosis and MDS. Panel A displays ROC for CMML when compared with reactive monocytosis yielding an AUC of 0.9592 (P < .001). Cut-off value for CM differentiating CMML from reactive monocytosis at 94.15% yielded sensitivity of 100% and specificity of 86.21%. Similarly, panel B displays ROC for CMML when compared with MDS. It yielded an AUC of 0.8779 (P < .001). Furthermore, the cut-off value for CM of 94.09%, yielded sensitivity of 71.76% and specificity of 86.21%.
ROC analysis of CMML in comparison with reactive monocytosis and MDS. Panel A displays ROC for CMML when compared with reactive monocytosis yielding an AUC of 0.9592 (P < .001). Cut-off value for CM differentiating CMML from reactive monocytosis at 94.15% yielded sensitivity of 100% and specificity of 86.21%. Similarly, panel B displays ROC for CMML when compared with MDS. It yielded an AUC of 0.8779 (P < .001). Furthermore, the cut-off value for CM of 94.09%, yielded sensitivity of 71.76% and specificity of 86.21%.
Additionally, no differences were seen when stratifying by lower and higher risk disease as defined by established cytogenetic risk stratification groups (P = .42) or by the prognostic scoring systems validated in CMML.5 Exposure to hypomethylating agent did not affect the pattern of CM in our cohort (P = .18), although this could be attributed to the lack of availability of sequential samples, both prior to treatment and at best response, from the same patient.
When comparing cases based on the presence of splicing mutations, DNA methylation mutation, ASXL1, or signaling mutations, no difference in classical monocyte pattern was identified. These observations collectively affirm that CMML can be accurately identified by the presence of CM irrespective of mutation status, the presence or absence of proliferative features, cytogenetic risk category, or overall prognostic classification.
Next, we sought to determine if CM was sufficient to discriminate pathologically defined CMML (n = 29) from MDS (n = 86) with no overlap in diagnosis. Indeed, CM was also capable of discriminating CMML from MDS with a sensitivity of 71.76% and a specificity of 86.21% (AUC, 0.8793; P < .0001) as depicted in Figure 1B. To explore the impact of CM in MDS, we stratified cases by the presence (n = 24) or absence (n = 62) of CM, based on our definition a priori. We compared pathologic, genetic, and clinical characteristics of “CMML-like” MDS, as defined by monocyte subset pattern. In this analysis, CMML-like MDS was associated with an increased WBC (3.815 × 109/L vs 2.34 × 109/L; P = .03), increased absolute neutrophil counts (1.73 × 109/L vs 1.07 × 109/L; P = .02), and increased absolute monocyte counts (AMC) (355 × 106/L vs 120 × 106/L; P = .02) compared with MDS cases with normal monocyte subsets (Figure 2). Furthermore, the MDS cohort without CM was more frequently associated with poor-risk cytogenetics (odds ratio [OR] 3.429; 95% confidence interval [CI], 1.032-10.08; P = .04) and was more likely to be higher risk as defined by the IPSS-R (OR, 8.77; 95% CI, 1.09-70.69; P = .02). Analysis of mutated genes identified SF3B1 to be present at greater frequency in the MDS subgroup with CM (OR, 3.46; 95% CI, 1.07-11.21; P = .04). The frequency of other commonly mutated genes in CMML or MDS were not significantly different between the 2 MDS cohorts (Figure 2). Although the significance of this finding needs further exploration, we report a higher number of cases associated with MDS ring sideroblasts and SF3B1 mutation in the MDS with CM cohort (20.8%) compared with the MDS without the CM cohort (8.33%). When comparing the overall survival, the MDS cohort with CM had a median survival of 83.3 months compared with 42.2 months in the MDS cohort without CM; however, it did not reach statistical significance (P = .0575).
Comparison of various clinical parameters in MDS cohort with presence or absence of CM. Total white count and AMC were more likely to be higher in the MDS cohort with CM when compared with the subgroup of MDS without presence of CM. Furthermore, patients in the MDS cohort who did not have CM were likely to have high-risk disease as determined by IPSS-R, and were likely to have poor-risk cytogenetics. SF3B1 mutations were found in greater frequency in the MDS cohort with presence of CM compared with the MDS subgroup with absence of CM (OR, 3.706; 95% CI, 1.117-12.26; P = .04). ALC, absolute lymphocyte count; ECOG, Eastern Cooperative Oncology Group; IPSS-R, revised International Prognostic Scoring System; PRBCs, packed red blood cells.
Comparison of various clinical parameters in MDS cohort with presence or absence of CM. Total white count and AMC were more likely to be higher in the MDS cohort with CM when compared with the subgroup of MDS without presence of CM. Furthermore, patients in the MDS cohort who did not have CM were likely to have high-risk disease as determined by IPSS-R, and were likely to have poor-risk cytogenetics. SF3B1 mutations were found in greater frequency in the MDS cohort with presence of CM compared with the MDS subgroup with absence of CM (OR, 3.706; 95% CI, 1.117-12.26; P = .04). ALC, absolute lymphocyte count; ECOG, Eastern Cooperative Oncology Group; IPSS-R, revised International Prognostic Scoring System; PRBCs, packed red blood cells.
Taken together, our study successfully validates and builds on the recently published study investigating the role of monocyte partitioning by multiparametric flow cytometry in CMML. Importantly, we validate the observation that CM can discriminate CMML cases from normal age-matched controls and MDS with high specificity and sensitivity, supporting its role in the diagnosis of CMML. We also demonstrated that MDS can be differentiated based on the presence of CM, and is clinically distinguished by a favorable prognosis and a higher frequency of SF3B1 mutations. Future investigations are warranted to understand the diagnostic, biologic, and clinical implications of CM in MDS.
Authorship
Contribution: E.P. designed the research; C.T. collected and analyzed the data; C.T. and E.P. wrote the paper; L.M., L.Z., and G.S. contributed to development and implementation of the monocyte subset flow cytometry test; A.K. contributed to data analysis and drafting of the manuscript; M.B. and Q.Z. contributed to data interpretation; R.K., A.F.L., K.S.Z., and J.E.L. contributed to data interpretation and drafting of the manuscript; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eric Padron, Department of Malignant Hematology, H. Lee Moffitt Cancer Center and Research Institute, 12902 Magnolia Dr, MRC3047A, Tampa, FL 33612; e-mail: eric.padron@moffitt.org.