To the editor:
Sézary syndrome (SS) is a rare, aggressive leukemic form of primary cutaneous T-cell lymphoma.1 The diagnostic criteria associate a clonal T-cell receptor (TCR) rearrangement with peripheral Sézary cell (SC) counts of ≥1 G/L, an increased CD4/CD8 ratio of ≥10, CD4+CD7− cells of ≥40%, or CD4+CD26− cells of ≥30%.2 The use of anti-TCR-Vβ antibodies provides a suitable tool for SCs, although 30% of the repertoire is not covered. We, and others, showed the specificity and the reliability of CD158k/KIR3DL2, a member of the killer-cell immunoglobulin-like receptor family, to identify blood-3-6 and skin-7,8 derived SCs. We developed an optimized flow cytometry strategy9 and reported that circulating CD158k+ T cells are more heterogeneous than expected, being not exclusively of CD62L+ CCR7+ central memory (TCM) T-cell phenotype.10-12
However, skin infiltration mechanisms and the relationships between circulating and infiltrating SCs are not fully understood.13 Here we characterize the phenotypic diversity of CD158k+ T cells in a large cohort of patients, including markers of recently described T-cell subsets, such as stem-cell memory (TSCM)14 and resident memory (TRM).15 A total of 47 SS patients diagnosed according to the International Society for Cutaneous Lymphomas– European Organization of Research and Treatment of Cancer classification2 were consecutively included in a prospective study at Saint-Louis Hospital. Blood- and skin-paired samples from 16 patients were also analyzed. The phenotypic diversity of SCs was further characterized at the transcriptomic level to provide molecular signatures of naïve/memory cell-sorted clones.
First, to investigate the heterogeneity of patients’ T cells, we characterized naïve (TN), TCM, transitional memory (TTM), effector memory (TEM), and terminal effector memory (TEMRA) subsets, on the basis of CD45RA, CCR7, CD27, and CD95 expression16 (supplemental Table 1, available on the Blood Web site). The recently identified TSCM subset, differing from TN by the expression of Fas receptor (CD95),14 possesses stem cell–like properties including self-renewal capacity and multipotency to generate memory subsets. The combination of anti-CD158k and TCR-Vβ labeling in 17 of 47 patients improved the discrimination between “clonal” Vβ+CD158k+ T cells and nonclonal Vβ−CD158k−CD4+ T cells (supplemental Figure 1A, supplemental Table 2).
Our results show that Vβ+CD158k+ T cells or CD158k+ T cells from 25 patients were not uniformly of the TCM phenotype, 18 of them expressing naïve markers (Figure 1A-B). Unexpectedly, 7 patients displayed Vβ+CD158k+ T cells or CD158k+ T cells of the TSCM phenotype, one having a homogeneous expansion of TSCM−CD158k+ T cells (Figure 1B, CERA45). TSCM cells are considered as the earliest memory T subset, and the concept of their stemness in cancer was first introduced in adult T-cell leukemia.17 Harnessing this potential may be valuable for the generation of cell lines or mouse models, which are poorly developed in SS. On the other hand, the expression of CD95, classically associated with memory CD45RA− subsets (healthy donor’s [HD’s] median 65%, 50% to 79%), was extremely heterogeneous among CD45RA−Vβ+CD158k+ T cells, from 0.6% to 97% (supplemental Figure 1B). A dysregulation of its expression on circulating SCs was already reported, up- or downregulation being involved in apoptosis resistance or tumorigenesis.18,19 During the follow-up, the initial naïve/memory phenotype of SC subsets fluctuated in 40% of the 29 patients monitored for more than 6 months, especially during blood relapse, as is shown for patient ARGP38, who shifted from TN to TCM (Figure 1B).
Phenotypic heterogeneity of circulating CD158k+T cells. (A) Among 47 patients, 28 display SCs of only 1 phenotype, mostly TCM, whereas 19 present a mix of several subsets, comprising naïve, TSCM, TCM, TTM, TEM, or TEMRA. (B) Example of 2 Sézary patients: CERA45, with 87% of SCs displaying a TSCM phenotype (CD45RA+CCR7+CD27+CD95+), stable over time; ARGP38, with SCs being exclusively naïve (85%) (CD45RA+CCR7+CD27+CD95−) in March 2014, presented a mix of naïve (66%) and TCM (30%) (CD45RA−CCR7+CD27+) in July 2016, along with an increase of blood tumoral burden and large-size lymphocytes (from 0.02 G/L CD158k+ T cells in November 2015 to 2.5 G/L in July 2016). (C) Vβ+CD158k+ T cells versus Vβ−CD158k−CD4+ T cells naïve/memory phenotype distribution within 17 patients. Ranges in 18 HDs are indicated by the blue backgrounds (mean ± 2 SD). Results were obtained after immunostaining on thawed PBMCs. Statistical analyses are paired among patients (Wilcoxon test), and a Mann-Whitney test was used for comparisons with HDs. Results are expressed as medians ± interquartile ranges. (D) Heat map resulting from supervised hierarchical clustering of differentially expressed genes among the TN, TSCM, and TCM phenotypes of CD4+ T cells from HDs and CD158k+CD4+ T cells from Sézary patients (P = .00001 | log2 fold change | > 1). ns, not statistically significant. *P < .05. **P < .01. ***P < .001.
Phenotypic heterogeneity of circulating CD158k+T cells. (A) Among 47 patients, 28 display SCs of only 1 phenotype, mostly TCM, whereas 19 present a mix of several subsets, comprising naïve, TSCM, TCM, TTM, TEM, or TEMRA. (B) Example of 2 Sézary patients: CERA45, with 87% of SCs displaying a TSCM phenotype (CD45RA+CCR7+CD27+CD95+), stable over time; ARGP38, with SCs being exclusively naïve (85%) (CD45RA+CCR7+CD27+CD95−) in March 2014, presented a mix of naïve (66%) and TCM (30%) (CD45RA−CCR7+CD27+) in July 2016, along with an increase of blood tumoral burden and large-size lymphocytes (from 0.02 G/L CD158k+ T cells in November 2015 to 2.5 G/L in July 2016). (C) Vβ+CD158k+ T cells versus Vβ−CD158k−CD4+ T cells naïve/memory phenotype distribution within 17 patients. Ranges in 18 HDs are indicated by the blue backgrounds (mean ± 2 SD). Results were obtained after immunostaining on thawed PBMCs. Statistical analyses are paired among patients (Wilcoxon test), and a Mann-Whitney test was used for comparisons with HDs. Results are expressed as medians ± interquartile ranges. (D) Heat map resulting from supervised hierarchical clustering of differentially expressed genes among the TN, TSCM, and TCM phenotypes of CD4+ T cells from HDs and CD158k+CD4+ T cells from Sézary patients (P = .00001 | log2 fold change | > 1). ns, not statistically significant. *P < .05. **P < .01. ***P < .001.
Using a subtractive gating strategy, excluding Vβ+CD158k+ T cells, we characterized the nonclonal TCD4+ subset in 17 patients (supplemental Table 2) with naïve/memory phenotype distribution similar to that of CD4+ T cells from HD (Figure 1C). Vβ+CD158k+ T cells were mostly but not fully CD26− (median 99%, range 6% to 100%). Strikingly, more than 30% of Vβ−CD158k−CD4+ T cells were CD26− in 14 patients (total patients’ median 44%, 8% to 74%) (supplemental Figure 1C-D). A similar trend for CD7 expression was observed, suggesting that T cells from SS patients display a skewed CD26− phenotype, CD7− phenotype, or both, not limited to SCs.
We then performed a transcriptomic analysis of sorted CD158k+CD4+ T cells from 3 patients, having a phenotype of TN, TSCM, TCM, or all 3. Of note, 2 types of clonal Vβ2+CD158k+ SCs (TCM and TSCM) were isolated for patient SYLN27. Results were confronted to sorted TN, TSCM, and TCM CD4+ T cells from 3 HDs. Unsupervised hierarchical clustering showed that all HD’s CD4+ T cells segregate into the same cluster and then according to their respective state of differentiation (supplemental Figure 2). By contrast, SCs displayed a distinct signature, which may be explained by genetic alterations involved in SS,20 such as overexpression of KIR3DL2, PLS3 (T-plastin), and TWIST1 (supplemental Figure 3). We then focused on transcripts discriminating naïve/memory subsets in the 3 HDs. Results in Figure 1D and supplemental Figure 4 showed that transcript profiles from SYLN27- and MARC54-sorted CD158k+ TCM are similar to those of HD’s TCM. More important, the second clonal Vβ2+CD158k+ subset from SYLN27, with phenotypic characteristics of TSCM, displayed molecular signatures close to those from HD’s TSCM. Of note, the naïve clonal CD158k+ subset from COIJ47 did not segregate with normal CD4+ TN but with TSCM. It can be noticed that SYLN27 TSCM and COIJ47 TN expressed naïve genes at a low level, with an atypical expression of some memory genes (supplemental Figure 4). According to their molecular signature, “naïve” SCs from COIJ47 may be reassigned as TSCM with an underexpression of FAS (supplemental Table 3), reflecting here a possible apoptosis resistance mechanism rather than a maturation feature.18
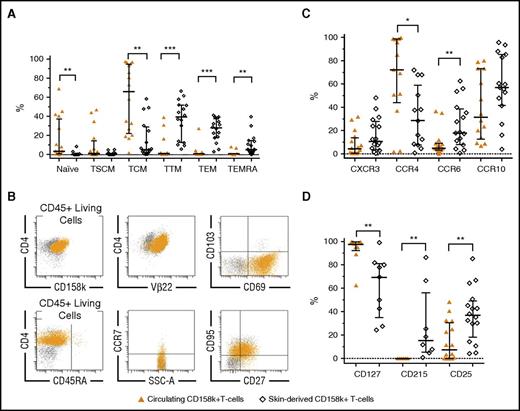
We next compared the distribution of naïve/memory SCs from blood and skin. Both compartments contained tumor cells with distinct differentiation profiles (Figure 2A; gating strategy in skin; supplemental Figure 5A). Skin-derived SCs, defined as CD158k+ T cells, Vβ+CD158k+ SCs, or both (supplemental Table 4), were exclusively memory (Figure 2B). According to the progressive model of T-cell differentiation,21 our data are in favor of “early” phenotypes in the blood and more “advanced” phenotypes in the skin, similar to skin CD4+ T cells from HDs (supplemental Table 5). The C-type lectin CD69, normally expressed by TRM,15,22 was quasi-exclusively found on skin-derived SCs (median 75%, range 44% to 90%, n = 7), in association or not with the CD103 integrin (Figure 2B; supplemental Figure 5B). These results suggest that a continuous recirculation of TCM-SCs between blood and skin may not be applicable to all SCs. In the light of these results, the relationships between circulating and infiltrating SCs need to be revisited.
Phenotypic discrepancies between circulating CD158k+T cells and skin-derived SCs. (A) Naïve/memory phenotypes, in favor of more mature subtypes in skin-derived SCs (n = 16). (B) Example of characterization of skin-derived Vβ22+CD158k+ T cells from patient MOTM35, showing the exclusively memory TTM/TEM phenotypes (CD45RA−CCR7−CD27±CD95+). These SCs express CD69, a marker of TRM, but not CD103. Ninety-four percent of the CD158k+ T cells coexpress TCRVβ22. (C) Expression CXCR3, CCR6, CCR10 (n = 16), and CCR4 (n = 13). (D) Expression of CD127 (IL-7Rα, n = 9), CD215 (IL-15Rα, n = 8), and CD25 (IL-2Rα, n = 16). Results were obtained from immunostaining on fresh whole blood and extracted cutaneous cells from 16 patients. Statistical analyses are paired among patients (Wilcoxon test). Medians ± interquartile ranges are indicated.
Phenotypic discrepancies between circulating CD158k+T cells and skin-derived SCs. (A) Naïve/memory phenotypes, in favor of more mature subtypes in skin-derived SCs (n = 16). (B) Example of characterization of skin-derived Vβ22+CD158k+ T cells from patient MOTM35, showing the exclusively memory TTM/TEM phenotypes (CD45RA−CCR7−CD27±CD95+). These SCs express CD69, a marker of TRM, but not CD103. Ninety-four percent of the CD158k+ T cells coexpress TCRVβ22. (C) Expression CXCR3, CCR6, CCR10 (n = 16), and CCR4 (n = 13). (D) Expression of CD127 (IL-7Rα, n = 9), CD215 (IL-15Rα, n = 8), and CD25 (IL-2Rα, n = 16). Results were obtained from immunostaining on fresh whole blood and extracted cutaneous cells from 16 patients. Statistical analyses are paired among patients (Wilcoxon test). Medians ± interquartile ranges are indicated.
Besides a remarkable disparity between blood- and skin-derived CD158k+ T cells, and apart from CCR4, the proportion of cells expressing the homing receptors CXCR3, CCR6, and CCR10 tended to be higher in the skin (Figure 2C). CCR4+CD158k+ T cells accounted from 2% to 100% of circulating SCs and were less represented within skin SCs. This may explain the results from a recent study evaluating mogamulizumab, an anti-CCR4 monoclonal antibody, showing that the response rate is 2 times less in the skin than in the blood.23 Heterogeneity between both disease sites was also found regarding receptors of T-cell homeostatic interleukins IL-2, IL-7, and IL-15 (Figure 2D), which are involved in SS pathogenesis,24 with a particular IL-7Rα overexpression on blood SCs.
In conclusion, we highlight the great diversity of SCs, in terms of naïve/memory maturation phenotype and molecular signature, as well as cytokine/chemokine receptor expression. These results question the dogma of SCs being exclusively of the TCM phenotype. Skin-derived SCs show a more advanced maturation pattern than do their circulating counterpart. Our data, aiming to clarify the origin and evolution of SCs during the disease and the relationships between skin and blood compartments, may potentially provide new therapeutic targets for tomorrow’s immunotherapies.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: M.R. is funded by “Médaille d’Or de l’Internat” (Assistance Publique–Hôpitaux de Paris). This work was supported by INSERM and Projet Emergent Cancéropôle PaSe. The authors thank the Technology Platform from the Institut Universitaire d'Hématologie for cell sorting and molecular biology.
Contribution: M.R. performed research, analyzed the data, and wrote the paper; M.D. analyzed transcriptomic data; C.R.-W. provided blood and skin samples and followed the patients; A.M.-C. performed TCR repertoire analyses; A.A. was in charge of molecular biology assays; G.M. performed analyses and participated in writing the manuscript; L.H. performed research; A.B., M.B., and A.T. designed research and participated in writing the manuscript; and H.M.-T. designed research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hélène Moins-Teisserenc, INSERM UMR-1160, Institut Universitaire d’Hématologie, Hôpital Saint Louis, 1 av. Claude Vellefaux, 75010 Paris, France; e-mail: helene.moins@univ-paris-diderot.fr.