Key Points
Candidate SNP associations with survival outcomes after URD transplant are most likely false-positive findings.
Over 85% of candidate SNPs are not linked to a biochemical function; of those that are, about half are not linked to the candidate gene.
Abstract
Multiple candidate gene-association studies of non-HLA single-nucleotide polymorphisms (SNPs) and outcomes after blood or marrow transplant (BMT) have been conducted. We identified 70 publications reporting 45 SNPs in 36 genes significantly associated with disease-related mortality, progression-free survival, transplant-related mortality, and/or overall survival after BMT. Replication and validation of these SNP associations were performed using DISCOVeRY-BMT (Determining the Influence of Susceptibility COnveying Variants Related to one-Year mortality after BMT), a well-powered genome-wide association study consisting of 2 cohorts, totaling 2888 BMT recipients with acute myeloid leukemia, acute lymphoblastic leukemia, or myelodysplastic syndrome, and their HLA-matched unrelated donors, reported to the Center for International Blood and Marrow Transplant Research. Gene-based tests were used to assess the aggregate effect of SNPs on outcome. None of the previously reported significant SNPs replicated at P < .05 in DISCOVeRY-BMT. Validation analyses showed association with one previously reported donor SNP at P < .05 and survival; more associations would be anticipated by chance alone. No gene-based tests were significant at P < .05. Functional annotation with publicly available data shows these candidate SNPs most likely do not have biochemical function; only 13% of candidate SNPs correlate with gene expression or are predicted to impact transcription factor binding. Of these, half do not impact the candidate gene of interest; the other half correlate with expression of multiple genes. These findings emphasize the peril of pursing candidate approaches and the importance of adequately powered tests of unbiased genome-wide associations with BMT clinical outcomes given the ultimate goal of improving patient outcomes.
Introduction
For over a decade, researchers have conducted candidate gene-association studies of patient survival outcomes after allogeneic blood or marrow transplant (BMT). The intent of these studies was to identify genetic variants outside of the HLA region that would increase knowledge about clinical management or serve as a potential target for novel therapeutics.1-70 The majority of these studies tested for associations in small data sets, ranging from a few dozen to a few hundred patients and donors, and included heterogeneous diseases (spanning benign to malignant hematological diseases), related donors (RDs) and/or unrelated donors (URDs) with various degrees of HLA matching, and patients treated across multiple decades, from the 1980s through early 2000s. We conducted the first adequately powered evaluation of these candidate single-nucleotide polymorphism (SNP) and gene hypotheses using typed and imputed data from an existing genome-wide association study (GWAS) named DISCOVeRY-BMT (Determining the Influence of Susceptibility COnveying Variants Related to one-Year mortality after BMT) to replicate or validate these published associations.71,72 In addition, we leveraged the available genome-wide data from DISCOVeRY-BMT and measured the aggregate association of all SNPs in the candidate genes with survival outcomes to determine how many of these candidate genes play a significant role in survival after transplant. Lastly, using publicly available data, we characterized the potential functionality of each candidate SNP in relation to the gene of interest.
Methods
Literature search
An extensive literature search of PubMed was performed to identify peer-reviewed scientific studies (published on or before 30 December 2016) that reported non-HLA genetic polymorphisms associated with survival outcomes after allogeneic BMT, including disease-related mortality (DRM), progression-free survival (PFS), transplant-related mortality (TRM), and/or overall survival (OS).1-70 The PubMed search terms, filtering approach, and link to all articles described herein are provided in the supplemental Methods (available on the Blood Web site).
Study population
Previously published candidate genes and SNPs were examined using data from an existing GWAS called DISCOVeRY-BMT.72 This GWAS analyzes the association of recipient survival following an URD BMT with non-HLA genetic variation in recipient and donor genomes.72,73 In addition, DISCOVeRY-BMT tests the joint effect of recipient and donor genetic variation, termed a recipient-donor (R-D) mismatch, using the absolute value of the difference in the number of minor alleles between recipient and donor at each SNP. For example, the R-D mismatch value at a given SNP where the recipient is homozygous major (0 minor alleles) and the donor is heterozygous (1 minor allele) would be the absolute value of 0-1 = 1. All patients included in DISCOVeRY-BMT provided informed consent for inclusion in the Center for International Blood and Marrow Transplant Research (CIBMTR) registry. The National Marrow Donor Program (NMDP) and Roswell Park Cancer Institute Institutional Review Boards approved the study protocol. Paired donor and recipient biospecimens and corresponding clinical data were obtained from the CIBMTR biorepository and database. DISCOVeRY-BMT consists of 2 independent cohorts of patients with acute myeloid leukemia (AML), acute lymphoblastic leukemia (ALL), or myelodysplastic syndrome (MDS). Patients were excluded if they received T-cell–depleted or cord blood grafts. Cohort 1 included 2499 10 of 10 HLA-matched URD-BMT R-D pairs with AML, ALL, or MDS from 2000 to 2008. Cohort 2 included 920 10 of 10 HLA-matched URD-BMT R-D pairs with AML, ALL, or MDS from 2009 to 2011 as well as 8 of 8 (but not 10 of 10) HLA-matched URD-BMT recipients from 2000 to 2011.72 Causes of death were adjudicated by an expert panel and the primary cause of death was defined as DRM or TRM.72
Genotyping and imputation
All samples were genotyped using the Illumina Human OmniExpress BeadChip and the Illumina HumanExome BeadChip (University of Southern California Genomics Facility). Samples were assigned to plates to ensure the even distribution of patient characteristics and potential confounding variables using Optimal Sample Assignment Tool (OSAT), an R/Bioconductor software package.74 Ninety percent of DISCOVeRY-BMT patients self-reported as European American, Caucasian, or white, thus replication and validation analyses were performed on these R-D pairs. Stringent quality control was done on both samples and SNPs within this population. Population outliers were removed using EIGENSTRAT75 (n = 73). Additional sample quality control removed samples with missing call rate ≥2% (n = 54), sex mismatch (n = 9), abnormal inbreeding coefficients (n = 20), and evidence of cryptic relatedness (n = 17), yielding 2111 and 777 recipients in cohorts 1 and 2, respectively. Typed SNPs were removed if the call rate was <98%, there was deviation from Hardy-Weinberg equilibrium proportions, or discordance between duplicate samples was >2%. In total, 637 655 and 632 823 SNPs from the OmniExpress BeadChip were available for imputation in cohorts 1 and 2, respectively, using 1000 Genomes Project phase 3. IMPUTE2 software was used for imputation and QCTOOL was used to remove imputed genotypes with an info score <0.7, certainty <0.7, and a minor allele frequency <0.005.76,77 The R-D mismatch genome dosage calculations, described in “Methods” under “Study population,” were done as the absolute value of the recipient minus donor minor allele dosages. Rs2066847 (SNP13) in NOD2/CARD15 was the only variant analyzed from the Illumina HumanExome as it was not typed on the OmniExpress chip or available following imputation.
Survival analyses
Prior to genetic analyses, clinical covariates for inclusion in genome-wide survival models were selected using bidirectional stepwise Cox proportional hazard models of OS, PFS, TRM, and DRM using R statistical software.78 Cox proportional hazard models of OS, TRM, and DRM evaluated SNPs associated with time to death with all survivors censored at 1 year post-BMT.79 PFS was defined as the time to disease progression or death. Deaths from TRM and DRM were treated as competing risks and analyzed accordingly.80 SNP models for OS adjusted for recipient age, disease status (early/intermediate or advanced), and graft source (blood or marrow); PFS and DRM SNP models adjusted for recipient age and disease status; TRM SNP models adjusted for recipient age, graft source, and body mass index (underweight/normal, overweight, or obese). Dosage data accounting for the probability of each genotype were used in all analyses of imputed data. Effect-size estimates and standard errors from DISCOVeRY-BMT cohorts 1 and 2 were compared and combined using a fixed-effects inverse variance meta-analyses in METAL. For each SNP, heterogeneity of effect-size estimates between cohorts 1 and 2 was assessed using P values from significance tests of heterogeneity (Phet) and I2.81 Variants with Phet < .05 and I2 > 50 were meta-analyzed with a random effects models using meta in R.82
Replication and validation of candidate gene studies
Results from genetic-association studies should be reproduced in independent samples in order to confirm findings.83 Researchers have defined 2 distinctive terms to describe the reproducibility based on differences between the original study population and the confirmation studies: replication and validation.84 Replication is defined as the original and confirmation studies both having similar inclusion criteria (including the same ethnic/ancestral population) so that any differences between the study populations can be attributed to random variation.84 Validation is defined as the original and confirmation study populations having different inclusion criteria (including different ethnic/ancestral populations) so that any differences between the original and confirmation study could be due to systematic variation.84 Thus, replication analyses were conducted when the original study included HLA-matched URD BMTs in patients of European ancestry. Validation analyses were performed on studies of leukemia patients of non-European ancestry, patient populations who received a BMT from a matched RD, or patient populations that were mixed between those who received a BMT from RDs and URDs. For studies of outcomes involving multiple hematologic malignancies, the entire DISCOVeRY-BMT study population was analyzed. If the original study population was specified as AML, ALL, and/or MDS, the same disease inclusion criteria were applied so that the replication/validation study population aligned with that of the original study population.
Gene-based association testing
VErsatile Gene-based Association Study 2 (VEGAS2) software was used for gene-based association testing.85 VEGAS2 uses 106 Monte Carlo simulations to test the global significance of an association for sets of SNPs in defined genomic regions. VEGAS2 reports a gene-based P value for each gene determined using individual SNP association P values. Directional effects are not incorporated into analyses; thus, all SNPs can be aggregated without dampening an association signal. For the gene-based replication or validation analyses, the P values from typed and imputed SNPs in DISCOVeRY-BMT (± a 10-kb flanking region) meta-analyses of OS, PFS, TRM, and DRM were used as input into the VEGAS2 software. Gene-based P values were calculated for donor, recipient, and R-D mismatch analyses of the full cohort (ALL, AML, and MDS patients) or homogenous disease subgroups (ALL or AML or MDS patients), corresponding to the analyses performed in the original studies.
Functional annotation
RegulomeDB,86 the Blood Expression Quantitative Trait Loci (eQTL) Browser,87 and Variant Effect Predictor (VEP)88 were used to provide functional annotation of the candidate SNPs. For each database, the raw data scores, P values, and annotations, respectively, were downloaded from each website and assigned to each SNP in our list. RegulomeDB scores are categorized as follows: 1a-1f are likely to affect transcription factor binding and linked to expression of a gene target; 2a-2c are likely to affect transcription factor binding; 3a-3b are less likely to affect transcription factor binding, and ≥3 has minimal binding evidence. A RegulomeDB score is assigned based on the level and evidence of functional modification attributable to the SNP86,89 in multiple cell lines from a range of tissues, with scores from 1 to 7, with 1 having the highest functional effect, supported by experimental evidence and 7 having no modifying effect.89 The RegulomeDB database derives these annotations using the publicly available data sets from Gene Expression Omnibus (GEO), the Encyclopedia of DNA Elements (ENCODE) project, and the Roadmap Epigenome Consortium. The Blood eQTL data are derived from a study of correlations between genetic variants and gene expression in over 5000 patients, with replication in almost 3000 individuals. Herein, we consider only cis-eQTLs, defined as <250 kb in distance between the SNP chromosomal position and the probe midpoint for gene expression. VEP was used to determine the hypothetical functional importance of missense and nonsense variants based on SIFT,90 Mutation Taster,91 and PolyPhen-2.92
Results
DISCOVeRY-BMT patient characteristics
DISCOVeRY-BMT cohorts 1 and 2 include mostly 10 of 10 HLA-matched URDs, with 281 8 of 8 HLA-matched donor-recipient pairs in cohort 2; all patients are of European continental ancestry. Cohorts do not differ by intensity of conditioning regimen, recipient or donor sex proportions, or Karnofsky performance score/Lansky performance status. However, cohort 1 includes more ALL patients whereas cohort 2 includes more recipients with MDS. AML disease status also differs between cohorts at P < .01 (Table 1).
Candidate gene studies of survival outcomes
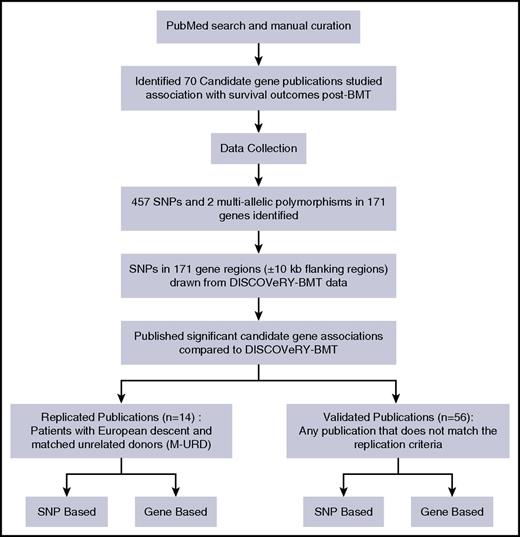
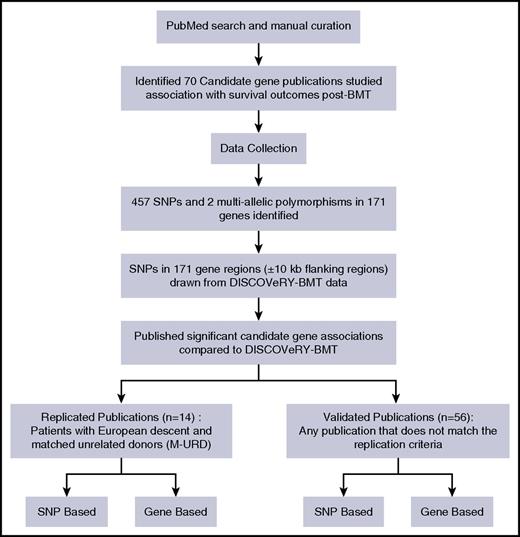
The literature search identified 70 publications that studied a total 458 SNPs and 2 multiallelic polymorphisms in 171 genes (Figure 1; supplemental Table 1). Studies included patients who received a transplant from an HLA-matched URD (19 articles), an HLA-matched RD (23 articles), or both (28 articles) (supplemental Table 1). Study populations included patients and donors of European ancestry (53 articles), Asian ancestry (15 articles), or mixed genomic ancestry (2 articles) (supplemental Table 1).
Pipeline performed to either replicate or validate candidate gene-association studies.
Pipeline performed to either replicate or validate candidate gene-association studies.
A total of 14 articles assessed genetic variation in HLA-matched URD BMT patients of European ancestry, but only 7 of these articles reported significant associations (P < .05 or an author-specified significance threshold) and thus comprise our replication study (supplemental Tables 2-3). A total of 56 articles tested associations in either a combination of RDs and URDs (RD-URD), only RD, and/or in non-European populations; 39 of these 56 articles reported at least 1 significant SNP association with survival outcome and we attempted to validate the significant findings from these 39 articles (supplemental Tables 2 and 4).
Replication
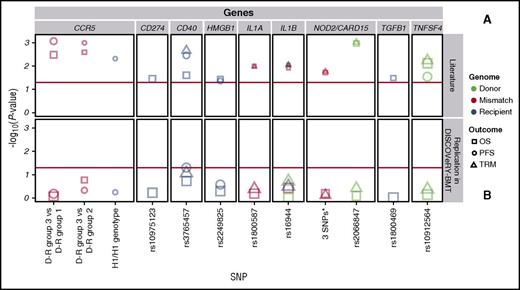
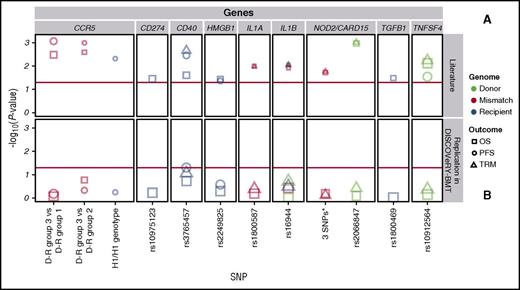
DISCOVeRY-BMT cohorts were used to replicate published studies of European American acute leukemia or MDS patients treated with an URD BMT.1-14 Of the 7 articles whose findings we attempted to replicate, 2 articles tested multiallelic models in NOD2/CARD155 and CCR56 ; 5 articles tested single SNP associations in TGFB1,1 CD274,3 CD40,3 TNFSF4,3 HMGB1,4 IL1A,7 IL1B,7 and NOD2/CARD152 (Table 2; Figure 2; supplemental Table 3).1-7
Replication attempts of previously reported significant candidate gene-association studies in DISCOVeRY-BMT. Survival association P values as reported in previous literature (A) and replication attempts of these associations in DISCOVeRY-BMT cohort (B) are shown as data points. Horizontal panels indicate the genes that these polymorphisms and haplotypes are located in or close to as reported by the previous studies. Shapes represent associations with survival outcomes OS, PFS, or TRM; colors correspond to donor, recipient, or donor-recipient mismatch polymorphisms. The size of the point represents the sample size of the study, with larger points reflecting a bigger sample size. Shown on x-axis are the 9 polymorphisms from the literature reporting associations at P < .05 with OS, PFS, or TRM by 1 or more previously published studies; the y-axis is the −log10 (P value). The red horizontal lines in (A) and (B) indicate P = .05. Details on the haplotypes are described in “Results” under “Replication.”
Replication attempts of previously reported significant candidate gene-association studies in DISCOVeRY-BMT. Survival association P values as reported in previous literature (A) and replication attempts of these associations in DISCOVeRY-BMT cohort (B) are shown as data points. Horizontal panels indicate the genes that these polymorphisms and haplotypes are located in or close to as reported by the previous studies. Shapes represent associations with survival outcomes OS, PFS, or TRM; colors correspond to donor, recipient, or donor-recipient mismatch polymorphisms. The size of the point represents the sample size of the study, with larger points reflecting a bigger sample size. Shown on x-axis are the 9 polymorphisms from the literature reporting associations at P < .05 with OS, PFS, or TRM by 1 or more previously published studies; the y-axis is the −log10 (P value). The red horizontal lines in (A) and (B) indicate P = .05. Details on the haplotypes are described in “Results” under “Replication.”
The 2 NOD2/CARD15 associations were based on a 3-variant R-D pair model (rs2066844 [SNP8], rs2066845 [SNP12], and rs2066847 [SNP13]) and single SNP associations with SNP13.27 The null type is when the R-D pair are the homozygous common allele for all 3 SNPs and the effect allele combination is the presence of 1 or more minor alleles at any of the 3 SNPs within the R-D pair. In a study of 196 patients who received an URD BMT for AML or ALL, the NOD2/CARD15 multi-SNP model was significantly associated with OS (relative risk [RR], 1.6; 95% confidence interval [CI], 1.1-2.4; P = .02) and TRM (RR, 1.6; 95% CI, 1.1-2.4; P = .02).5 However, in the DISCOVeRY-BMT AML and ALL patients (n = 1597) treated with an URD BMT, there was no association with OS (hazard ratio [HR], 1.03; 95% CI, 0.9-1.2; P = .72) or TRM (HR, 1.1; 95% CI, 0.8-1.4; P = .6) (Figure 2; supplemental Table 3). In a study of 342 URD genotypes matched with AML or ALL patients, rs2066847 (SNP13) alone significantly increased risk of TRM and OS approximately threefold (P = .001) and 2.5-fold (P = .001), respectively2 ; however, DISCOVeRY-BMT donor genotypes did not associate with either TRM (HR, 1.17; 95% CI, 0.78-1.74; P = .45) or OS (HR, 0.98; 95% CI, 0.73-1.31; P = .89) in ALL or AML patients (Table 2; Figure 2; supplemental Table 3).
One of the largest candidate gene studies (N = 1370) showed significant associations between PFS and recipient CCR5 H1/H1 genotype (n = 163), as well as with author-defined genotype risk subgroups and OS.6 In DISCOVeRY-BMT, neither the CCR5 H1/H1 genotype (n = 294) nor the genotype risk groups defined by H1/H16 status were significantly associated with PFS or OS (Figure 2; supplemental Table 3). The genotype risk groups tested by the authors were substantially smaller than the full cohort (Table 2). In DISCOVeRY-BMT, these subgroups were approximately twice as large as those in the original study and adequately powered to detect these associations. Attempts to replicate single SNP associations in TNFSF4,3 TGFB1,1 HMGB1,5 IL1A,7 and IL1B7 also failed (Table 2; Figure 2; supplemental Table 3).
Validation
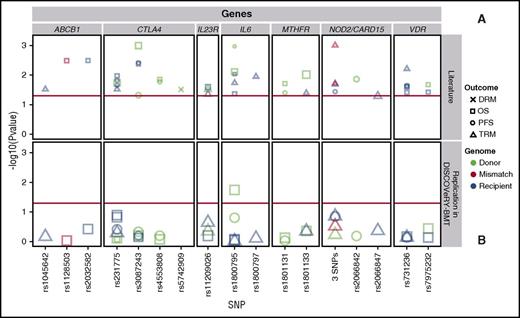
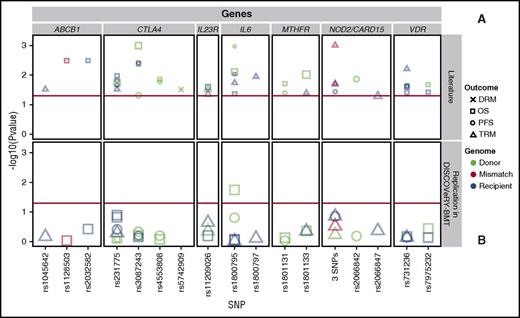
We attempted to validate 36 polymorphisms in 26 genes from 39 candidate gene articles (supplemental Tables 2 and 4),15-52 including: ABCB1,29,32 CD14,42 CTLA4,28,40,43-46,51 CYP2C19,38 DAAM2,52 EP300,36 ESR1,17 GSTA2,19 GZMB,24 ICAM1,48 IL23R,20,22 IL6,15-17 IRF3,37 KLRK1,23 LIG3,48 MTHFR,31,35,41 MUTYH,48 NOD2/CARD15,25,27,30,33,50 NOS1,30 P2RX7,34 TDG,48 TIRAP,17 TLR4,42 TYMP,26 and VDR.18,21,39,47 These studies reported significant genetic associations with survival after transplant in patients who received a HLA-matched RD BMT (19 articles) or had a study population including HLA-matched RD and URD BMT patients, without stratification of results (17 articles). We also attempted to validate survival associations seen in non-European leukemia patients who received an URD BMT (3 articles). We present results of variants reported significant in at least 2 separate publications in Table 3 and Figure 3.
Validation attempts of previously reported significant candidate gene-association studies in DISCOVeRY-BMT at least twice. Survival association P values as reported in previous literature (A) and validation attempts of these associations in DISCOVeRY-BMT cohort (B) are shown as data points. Horizontal panels indicate the genes that these 17 polymorphisms are located in or closest to as reported by the previous studies. Shapes represent associations with survival outcomes DRM, OS, PFS, and TRM; colors correspond to donor, recipient, or donor-recipient mismatch polymorphisms. The size of the point represents the sample size of the study, with larger points reflecting a bigger sample size. Shown on x-axis are the 17 polymorphisms from the literature reporting associations at P < .05 with OS, PFS, or TRM by 1 or more previously published studies; the y-axis is the −log10 (P value). The red horizontal lines in (A) and (B) indicate P = .05. Details on the haplotypes are described in “Results” under “Validation.”
Validation attempts of previously reported significant candidate gene-association studies in DISCOVeRY-BMT at least twice. Survival association P values as reported in previous literature (A) and validation attempts of these associations in DISCOVeRY-BMT cohort (B) are shown as data points. Horizontal panels indicate the genes that these 17 polymorphisms are located in or closest to as reported by the previous studies. Shapes represent associations with survival outcomes DRM, OS, PFS, and TRM; colors correspond to donor, recipient, or donor-recipient mismatch polymorphisms. The size of the point represents the sample size of the study, with larger points reflecting a bigger sample size. Shown on x-axis are the 17 polymorphisms from the literature reporting associations at P < .05 with OS, PFS, or TRM by 1 or more previously published studies; the y-axis is the −log10 (P value). The red horizontal lines in (A) and (B) indicate P = .05. Details on the haplotypes are described in “Results” under “Validation.”
Our validation analyses identified only 1 variant associated at P < .05. Donor variation in rs1800795 (IL-6) associated with OS (HR, 1.11; 95% CI, 1.0-1.2; P = .02) (Figure 3; supplemental Table 4). This SNP association was initially reported in a single study by Balavarca et al17 (HR, 1.29; 95% CI, 1.07-1.55; P = .007) in patients with acute leukemia, CML, or lymphoma treated with a matched RD or URD BMT (n = 743).
SNPs within NOD2/CARD15 were the most frequently studied and reported of all candidate gene-association studies in our validation set (supplemental Table 2). NOD2/CARD15 is a susceptibility gene for inflammatory bowel disease and may be involved in Crohn disease.27 We attempted to validate studies that reported an association of NOD2/CARD15 and survival outcomes in HLA-matched RD and URD BMT patients27,30,33 or HLA-matched RD BMT patients.25,50 Three studies reported significant findings between the presence of the NOD2/CARD15 multi-SNP polymorphism in either donor or recipient with TRM27,50 or PFS,25 however, this did not validate in the DISCOVeRY-BMT cohorts (Figure 3; Table 3). There was also no significant association of the single variant rs2066842 in RDs/URDs with PFS,30 or the single variant rs2066847 (SNP13) in recipients of RD/URD BMTs with TRM (Figure 3; Table 3)33 in the DISCOVeRY-BMT cohorts.
Due to its known functions and perceived implications in transplant biology,43 associations with multiple SNPs in CTLA4 have been tested in numerous transplant populations (supplemental Table 2), with 4 CTLA4 SNPs (rs3087243, rs231775, rs4553808, rs5742909) reported as significantly associated with survival after RD or URD allogeneic BMT in acute leukemias, CML, lymphomas, MDS, and other hematological disorders (Table 3). Attempts to validate CTLA4 SNPs with DRM, PFS, OS, and TRM were unsuccessful in the DISCOVeRY-BMT cohorts (Table 3; Figure 3; supplemental Table 4).
The remaining results of the 25 additional candidate genes containing SNPs that were tested in the DISCOVeRY-BMT cohorts are summarized in supplemental Tables 4 and 3 as well as Figure 3; no SNP associations were found at P < .05. Importantly, the P value distribution of the single SNP associations showed no deviation from the null expectation with 95% CIs (supplemental Figure 2), suggesting that we cannot reject the null hypothesis of no association with survival outcome.
Gene-based replication and validation of previous studies
The reviewed candidate gene studies first selected genes based on their hypothesized or known function, and subsequently selected variants within that gene for single SNP or haplotype testing. Thus, although SNPs and haplotypes were tested individually for association, the hypotheses from the literature can be considered gene-based. The density of typed and imputed markers in the DISCOVeRY-BMT recipients and donors allows us to measure the aggregate effect of all SNPs within each candidate gene on survival. Genes were selected for testing from the same literature used to perform the replication and validation SNP and haplotype analyses. VEGAS2 gene-based testing did not reveal any associations at P < .05 with any of the survival outcomes in either the replication or validation groups (supplemental Table 5).
Candidate polymorphism annotation
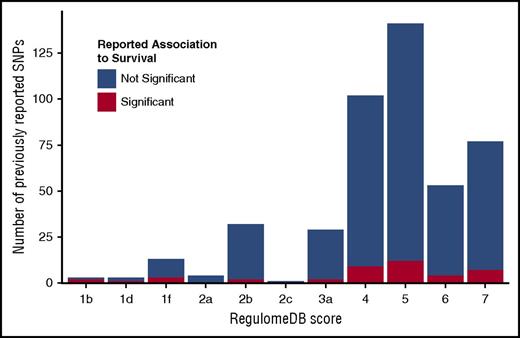
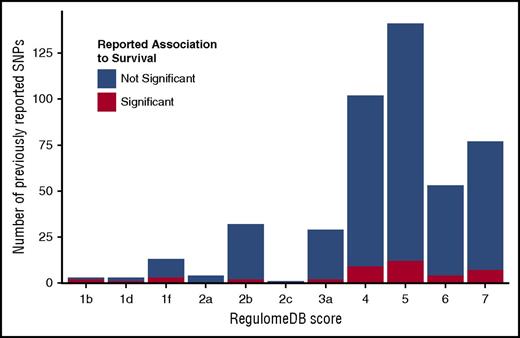
Candidate gene SNPs were analyzed using the RegulomeDB,86 VEP,88 and Blood eQTL Browser87 databases to assess their functional characteristics and better understand their biological framework. Eighty percent of previously reported SNPs had RegulomeDB scores >3 (Figure 4; supplemental Table 6), indicating that these SNPs have minimal to no effect on modifying transcription. This distribution aligns with the overall distribution of SNPs in the genome, thus the candidate SNPS are not enriched for their impact on gene expression or transcription factor binding. Our replication and validation analyses include 2 protein-coding variants; VEP shows that only rs2066845 (SNP12) in NOD2/CARD15 is predicted to be damaging and disease causing.
RegulomeDB score distribution of previously studied polymorphisms. RegulomeDB categories are shown on the x-axis; counts of SNPs falling into RegulomeDB score category are shown on the y-axis. Blue portion of the bar indicates the counts of SNPs that were tested but not reported significant; red portion shows the counts of SNPs that were reported significant at least once. Score descriptions are given below the image. 1b indicates eQTL + transcription factor (TF) binding + any motif + DNase footprint + DNase peak; 1d, eQTL + TF binding + any motif + DNase peak; 1f, eQTL + TF binding/DNase peak; 2a, TF binding + matched TF motif + matched DNase footprint + DNase peak; 2b, TF binding + any motif + DNase footprint + DNase peak; 2c, TF binding + matched TF motif + DNase peak; 3a, TF binding + any motif + DNase peak; 4, TF binding + DNase peak; 5, TF binding or DNase peak; 6, motif hit; 7, no evidence.
RegulomeDB score distribution of previously studied polymorphisms. RegulomeDB categories are shown on the x-axis; counts of SNPs falling into RegulomeDB score category are shown on the y-axis. Blue portion of the bar indicates the counts of SNPs that were tested but not reported significant; red portion shows the counts of SNPs that were reported significant at least once. Score descriptions are given below the image. 1b indicates eQTL + transcription factor (TF) binding + any motif + DNase footprint + DNase peak; 1d, eQTL + TF binding + any motif + DNase peak; 1f, eQTL + TF binding/DNase peak; 2a, TF binding + matched TF motif + matched DNase footprint + DNase peak; 2b, TF binding + any motif + DNase footprint + DNase peak; 2c, TF binding + matched TF motif + DNase peak; 3a, TF binding + any motif + DNase peak; 4, TF binding + DNase peak; 5, TF binding or DNase peak; 6, motif hit; 7, no evidence.
The Blood eQTL Browser determines whether candidate SNPs have a significant role in cis gene expression of the candidate gene. Of the 171 genes included in our literature search results, 52% have at least 1 significant cis-eQTL at a probe-level false discovery rate (FDR) < 0.05. On a genome-wide level, ∼44% of genes have blood cis-eQTLs (FDR, P < .05). However, despite over half of the candidate genes having blood cis-eQTLS, only 13% of the candidate SNPs reported in these articles are blood cis-eQTLs. Thus, although blood eQTLs have been identified in these genes, they were not genotyped and analyzed in these candidate gene studies. Furthermore, almost half of the eQTLs in the candidate gene studies are correlated with expression that is not the candidate gene but rather a nearby gene. For example, rs7975232 (VDR) is an eQTL for SLC48A1 whereas the CTLA4 SNPs are actually eQTLs for CD28. The remaining eQTLs were correlated with expression of the candidate gene of interest, but in most cases, were also significant eQTLs for several other nearby genes (supplemental Table 6).
Discussion
Our study aimed to replicate or validate all previous genetic-association studies that investigated the non-HLA genetic effects on allogeneic BMT survival. Because previous studies selected SNPs in candidate genes, we conducted both single SNP and gene-based analyses to determine the aggregated SNP associations within candidate genes while still accounting for dependence between signals due to linkage disequilibrium.
The only association with P < .05 in our replication and validation analyses using DISCOVeRY-BMT was the donor SNP rs1800795 in IL-6 with OS. As reported,18 the rationale for studying this SNP was based on the immunological function of IL-6 and 2 prior findings showing that it was associated with graft-versus-host disease (GVHD),93 and response to chronic hepatitis C virus therapy.94 We found no evidence of association at P < .05 between donor SNP rs1800795 with death due to either GVHD or infection in the DISCOVeRY-BMT cohort (data not shown). Furthermore, rs1800795 is located in the intronic region of IL-6, has no effect on IL-6 expression or levels,95 but rather is an eQTL for 2 other nearby genes.95,96
In addition to exploring this IL-6 association further, we felt the validation of the CCR5 associations of H1/H1 genotype with outcome required additional efforts, as these associations were found in the largest study we attempted to validate; samples were also from CIBMTR (earlier years than our study population) and, unlike many of the other studies, survival effects only started to appear ∼2 years posttransplant. Analyses outlined in Table 2 were performed without censor at 1 year for OS (median survival time, 13.7 months; range, <1 to 125.6 months) and PFS (median time, 11.1 months; range, <1 to 125.6 months). There were no genotype associations with either outcome at P < .10.
Another frequently studied gene, CTLA4, highlights the heterogeneity specific to studies of genetic variation in transplant and perhaps helps explain why we did not replicate or validate associations. rs5742909 in CTLA4 was tested for association with various survival outcomes after transplant in 6 independent studies of HLA-matched-related donor-recipient pairs. In donors, the variant was found to be associated with DRM in 1 small study (N = 120), this was the only study that tested donor genotype with DRM. Likewise, 1 of 9 papers testing the association of rs231775 with survival outcomes measured the association of PFS with recipient rs231774 in 164 recipients (P = .025). Despite the frequency with which these 2 CTLA4 variants were studied, for both SNP-outcome combinations, DISCOVeRY-BMT is the only validation attempt. These SNPs are like those of many candidate gene hypotheses, in that they have not been tested in the same genome for the same outcome in similar populations, and if they have the N is small (supplemental Table 1).
Our inability to replicate or validate previous candidate gene associations could also be due to differences in inclusion criteria with respect to disease, donor relation, or to differences in our end point of 1-year survival vs longer-term survival. The previous genetic associations were hypothesized to be independent of underlying hematologic disease, therefore we would expect to replicate or validate these associations in a homogeneous patient population such as DISCOVeRY-BMT. When possible, we aligned our study population to the original candidate gene study (ie, restricted to AML patients only). Although DISCOVeRY-BMT focused on early 1-year survival, which may have different genetic contributions than later survival, many of the survival curves in the significant candidate gene articles show separation by genotype well before 1 year posttransplant, thus the significant published variants do not appear to correlated with only longer-term survival.
The large sample size of the DISCOVeRY-BMT provides adequate statistical power to attempt replication and validation of previously published candidate gene analyses,71 however, we did not reproduce these findings, similar to 2 other recent studies attempting to replicate previous candidate gene associations with GVHD after BMT.73,97 Other reports have also concluded that a substantial amount of the published candidate gene literature has presented false-positive associations.98
Confirming genetic-association studies is vital to identify true positive genetic variants that may contribute to complex phenotypes. False associations lead to wasted time, energy, and money in pursuit of confirmatory studies and could harm patients by delaying clinical discovery or by applying clinical studies too quickly without replication. Annotation of the previously reported SNP associations using publicly available data shows that few variants are functional; only 1 SNP is predicted to be damaging or deleterious, a small proportion of SNPs are correlated with gene expression, and an even smaller number are cis-eQTLs for the target gene of interest. Thus, although we did not replicate or validate these associations, the SNPs selected are not linked to functional annotation nor are they clearly related to the candidate genes. This underscores a fundamental problem with candidate gene studies that are hostage to the state of scientific knowledge at the time. Adequately powered testing of genetic associations with transplant outcomes remains critical to discovery and replication of genetic associations with the ultimate goal of improving patient outcomes.
Presented in part at the 58th annual meeting of the American Society of Hematology, San Diego, CA, 5-8 December 2016.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health (NIH), National Cancer Institute grant R03 CA188733 (L.E.S.-C. and T.H.). DISCOVeRY-BMT was supported by NIH, National Heart, Lung, and Blood Institute grant R01 HL102278 (L.E.S.-C. and T.H.). The Roswell Park Cancer Institute Biostatistics & Bioinformatics Core was partially supported by NIH, National Cancer Institute grant P30 CA016056. The Center for International Blood and Marrow Transplant Research was supported by Public Health Service grant/Cooperative Agreement 5U24-CA076518 from the NIH, National Cancer Institute, the NIH, National Heart, Lung, and Blood Institute, and the NIH, National Institute of Allergy and Infectious Diseases; grant/Cooperative Agreement 5U10HL069294 from the NIH, National Heart, Lung, and Blood Institute and the NIH, National Cancer Institute; contract HHSH250201200016C with Health Resources and Services Administration of the Department of Health and Human Services; grants N00014-15-1-0848 and N00014-16-1-2020 from the Office of Naval Research; and grants from Alexion; Amgen, Inc*; anonymous donation to the Medical College of Wisconsin; Astellas Pharma US; AstraZeneca; Be the Match Foundation; Bluebird Bio, Inc*; Bristol-Myers Squibb Oncology*; Celgene Corporation*; Cellular Dynamics International, Inc; Chimerix, Inc*; Fred Hutchinson Cancer Research Center; Gamida Cell Ltd; Genentech, Inc; Genzyme Corporation; Gilead Sciences, Inc*; Health Research, Inc Roswell Park Cancer Institute; HistoGenetics, Inc; Incyte Corporation; Janssen Scientific Affairs, LLC; Jazz Pharmaceuticals, Inc*; Jeff Gordon Children’s Foundation; The Leukemia & Lymphoma Society; Medac, GmbH; MedImmune; The Medical College of Wisconsin; Merck & Co, Inc*; Mesoblast; MesoScale Diagnostics, Inc; Miltenyi Biotec, Inc*; National Marrow Donor Program; Neovii Biotech NA, Inc; Novartis Pharmaceuticals Corporation; Onyx Pharmaceuticals; Optum Healthcare Solutions, Inc; Otsuka America Pharmaceutical, Inc; Otsuka Pharmaceutical Co, Ltd (Japan); PCORI; Perkin Elmer, Inc; Pfizer, Inc; Sanofi US*; Seattle Genetics*; Spectrum Pharmaceuticals, Inc*; St. Baldrick’s Foundation; Sunesis Pharmaceuticals, Inc*; Swedish Orphan Biovitrum, Inc; Takeda Oncology; Telomere Diagnostics, Inc; University of Minnesota; and Wellpoint, Inc* (*corporate members).
The views expressed in this article do not reflect the official policy or position of the National Institutes of Health, the Department of the Navy, the Department of Defense, Health Resources and Services Administration, or any other agency of the US government.
Authorship
Contribution: A.A.R. and E.K. performed research, analyzed and interpreted data, generated figures, and wrote the paper; L.M.P. analyzed and interpreted data; C.A.H., D.O.S., L.P., and X.S. performed genotyping interpretation of data; S.S. and M.C.P. acquired data and interpreted data analyses; X.Z. performed quality control and coding; P.L.M., K.O., A.I.C.-G., and D.T. interpreted data analyses; A.W., G.B., and S.B. performed quality control and coding; Q.Z., L.Y., Q.L., Q.H., and S.L. provided quality control and data randomization; T.H. conceived and designed the study, acquired data, and interpreted data analyses; L.E.S.-C. performed research, conceived and designed the study, acquired, analyzed, and interpreted data, and wrote the paper; and all authors participated in the revising of the manuscript, contributed critically important intellectual content, and gave approval of the final submitted version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lara E. Sucheston-Campbell, The Ohio State University, 496 W. 12th Ave, 604 Riffe Building, Columbus, OH 43210; e-mail: sucheston-campbell.1@osu.edu; and Theresa Hahn, Roswell Park Cancer Institute, Elm and Carlton Sts, Buffalo, NY 14263; e-mail: theresa.hahn@roswellpark.org.
References
Author notes
E.K. and A.A.R. contributed equally.
T.H. and L.E.S.-C. contributed equally.