Abstract
Introduction: Acute myeloid leukemia (AML) is the second most common pediatric leukemia with the worst prognosis of all major cancers in pediatrics. Resistance is the most common reason for treatment failure and adverse side effects also contribute to morbidity and mortality. Since cytarabine is the mainstay of AML treatment, the wide interpatient variability in response to cytarabine complicates chemotherapy further. This necessitates the identification of additional genetic variants important in the susceptibility to cytarabine cytotoxicity in order to allow for personalized treatment that maximizes efficacy and minimizes toxicity. Previous studies have utilized genome-wide gene expression signatures to identify markers associated with cytarabine response. Few studies have also utilized lymphoblastoid cell lines from normal individuals available through HapMap project to perform genome-wide association analysis (GWAS) to identify genetic-variants and gene-expression associated with in vitro cellular cytotoxicity to cytarabine. However, to best of our knowledge no GWAS study has so far been performed on in vitro cytarabine chemosensitivity of leukemic cells obtained from pediatric AML patients.
Hypothesis: To identify SNPs predictive of cytarabine in vitro chemosensitivityof leukemic cells obtained from pediatric AML patients (St Jude AML02 clinical trial) at diagnosis.
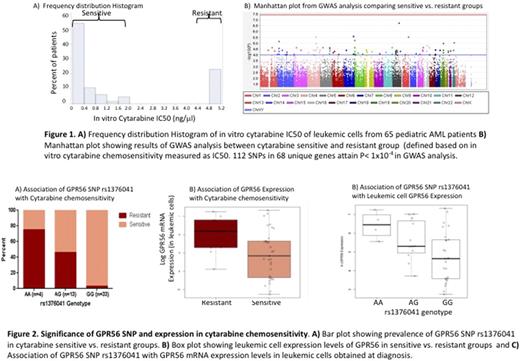
Methods: Leukemic cells from diagnosis obtained from N=65 patients were treated with varying concentrations of cytarabine and IC50 values were obtained.Samples were divided into resistant (n= 15; IC50 > 5ng/μl) and sensitive groups (n=50; IC50 < 5ng/μl), Figure 1A. Genome-wide SNPdata were generated using Illumina Omni 2.5M and Exome Beadchip SNP array at University of Miami, Hussman Institute for Human Genomics. We performed the standard GWAS QC procedure in order to remove SNPs with call rate < 95%, monomorphic SNPs, SNPs with MAF < 5% and samples with call rate < 95%. Following QC, GWAS was performed on the remaining high quality SNPs (n=1,317,146) to identify if any of these are significantly associated with resistant or sensitive groups. Second, genes to which SNPs passing the suggestive threshold (p < 1x10-4) map and genes within +/-500kb of these SNPs were tested for differential expression between sensitive and resistant groups using T-test. Third, SNPs mapping to genes or SNPs within 500kb from genes that were differentially expressed (nominal p<0.05) were then tested for their eQTL effect using T-test or ANOVA.
Results: Although none of the SNPs reached genome-wide significance (p<5.0 × 10-8), 112 SNPs located within or near 68 unique genes reached the suggestive level (p<1x10-4,Figure 1B). Among the top candidate SNPs were GPR56 rs1376041 G>A and rs75400242 G>ASNP found within 500kb of IGF1R gene. For both these SNPs the minor allele A was significantly associated with cytarabine resistance (p=1.42x10-5 and p=2.19x10-5, respectively). Furthermore, the minor allele of rs1376041 and rs75400242 was associated with a significantly higher level of GPR56 and IGF1R gene expression level (p=0.019 and p=0.044). Results for GPR56 for association of rs1376041 SNP with cytarabine resistance and GPR56 gene expression are shown in Figure 2A and 2C. Consistent with above observation for both GPR56 (Figure 2B) and IGF1R higher leukemic cell gene expression was associated with resistant group as compared to sensitive cases (p=0.0313 and p=0.0270).
Conclusions: These results suggest that these SNPs might contribute to cytarabine in vitro sensitivity through regulation of gene expression. This is biologically relevant since it has been documented that GPR56 expression is associated with cytarabine resistance and poor outcomes in AML. In fact, recent data suggests high expression of GPR56 to contribute to AML development identified it as a potential target for antibody targeted therapy. For IGF1R, consistent with our results it has been associated with promoting growth of AML cells and cytarabine resistance as well. Future functional pharmacogenomic studies will help in better understanding the mechanisms behind the SNP associations reported here.
Acknowledgments: We are thankful for funding from NIH R01CA139246 and Dr. McDonough for helpful discussions regarding GWAS analysis.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.