In this issue of Blood, Khoo et al of the Croucher/Phan group take advantage of a widely used mouse model of human multiple myeloma to define a molecular signature of tumor dormancy1 that is induced by interaction of myeloma cells with osteoblasts in the endosteal bone marrow niche2 ; is characterized by upregulation of myeloid genes, including Axl, in quiescent myeloma cells that retain the ability to spawn tumors upon reactivation of proliferation; and is amenable to pharmacological targeting using small-molecule AXL inhibitors.3
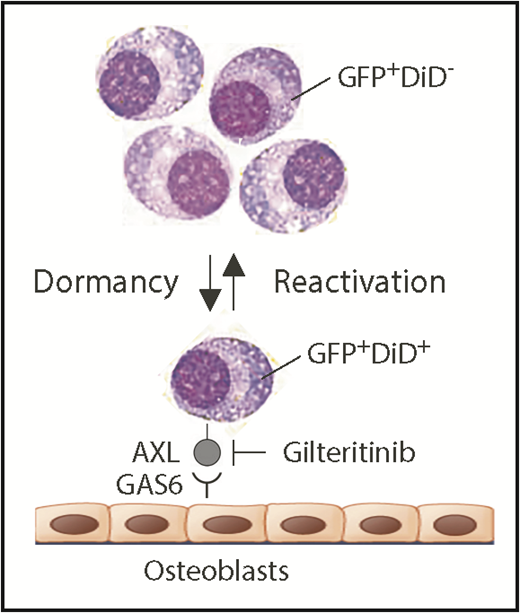
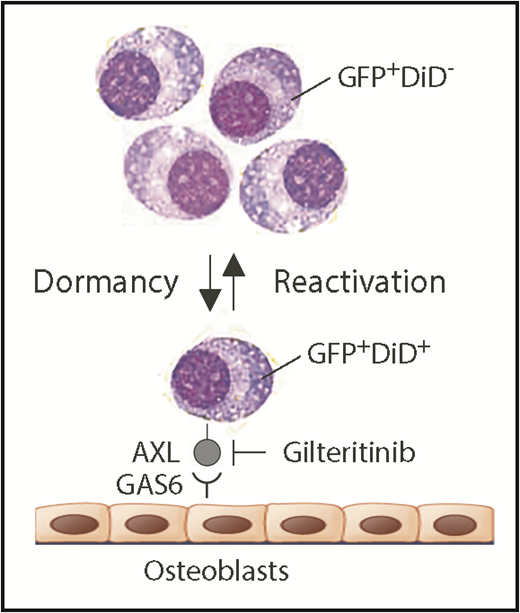
Schematic depiction of myeloma dormancy as a reversible, dynamic state induced, in part, by binding of GAS6 on osteoblasts to the receptor tyrosine kinase, AXL, on mouse 5TGM1 myeloma cells. In this experimental model system of human myeloma, the reactivation of a miniscule fraction of dormant tumor cells suffices to generate bulk myeloma in the hematopoietic bone marrow (eg, as few as ∼15 cells account for the total tumor burden in long bones of untreated mice).5 New findings reported implicate the endosteal niche in a novel, environmentally induced phenotypic change in dormant myeloma cells that includes the ectopic expression of myeloid genes, such as Axl and Vcam1 (which partners with α4β1 integrin on osteoblasts; not shown). Additionally, Khoo et al demonstrate that dormant myeloma cells can be dislodged from survival-protecting niches using small-compound AXL inhibitors. Gilteritinib, an AXL-inhibiting drug that very recently won US Food and Drug Administration approval for the treatment of adult patients with acute myeloid leukemia that harbors mutated FLT3, may lend itself to evaluating this possibility in clinical trials of myeloma. In addition to promoting dormant myeloma cells, the interaction of AXL and GAS6 in the osteoblastic niche also governs dormancy of prostate cancer cells9 and survival and self-renewal of chronic myeloid leukemia stem cells.10 GAS6, growth arrest specific 6.
Schematic depiction of myeloma dormancy as a reversible, dynamic state induced, in part, by binding of GAS6 on osteoblasts to the receptor tyrosine kinase, AXL, on mouse 5TGM1 myeloma cells. In this experimental model system of human myeloma, the reactivation of a miniscule fraction of dormant tumor cells suffices to generate bulk myeloma in the hematopoietic bone marrow (eg, as few as ∼15 cells account for the total tumor burden in long bones of untreated mice).5 New findings reported implicate the endosteal niche in a novel, environmentally induced phenotypic change in dormant myeloma cells that includes the ectopic expression of myeloid genes, such as Axl and Vcam1 (which partners with α4β1 integrin on osteoblasts; not shown). Additionally, Khoo et al demonstrate that dormant myeloma cells can be dislodged from survival-protecting niches using small-compound AXL inhibitors. Gilteritinib, an AXL-inhibiting drug that very recently won US Food and Drug Administration approval for the treatment of adult patients with acute myeloid leukemia that harbors mutated FLT3, may lend itself to evaluating this possibility in clinical trials of myeloma. In addition to promoting dormant myeloma cells, the interaction of AXL and GAS6 in the osteoblastic niche also governs dormancy of prostate cancer cells9 and survival and self-renewal of chronic myeloid leukemia stem cells.10 GAS6, growth arrest specific 6.
Owing to both newly developed myeloma drugs and the continuous refinement of therapeutic regimens that combine high-dose chemotherapy with autologous hematopoietic stem cell transplantation, the outcome for patients with multiple myeloma has significantly improved in recent years,2 making it possible at long last to consider a tangible number of patients as de facto cured.3 However, in the great majority of cases, following a period of successful therapy, myeloma relapses as a drug-refractory aggressive disease that leaves few, if any, therapeutic options. A large body of evidence indicates that a small but therapy-resistant subpopulation of tumor cells is responsible for disease relapse, thus composing the principal roadblock to curing myeloma. Depending on the investigator team, these cells are variably referred to as myeloma-initiating, myeloma-propagating, transit-amplifying, or cancer stem cell–like myeloma cells or, to use the crisp term preferred by Khoo et al, as dormant myeloma cells. The tight association of myeloma dormancy with minimal residual disease, acquired drug resistance, and tumor relapse4 underscores the urgent need to enhance our understanding of the biology and genetics of myeloma dormancy and to identify molecular targets for new approaches to kill “myeloma sleeper agents” in survival-protecting niches in situ or flush them out into the bone marrow and thereby resensitize them to chemotherapeutic attack.
Understanding that the research objective described is difficult to pursue in patients with myeloma, Khoo et al decided to use the well-established 5TGM1 mouse-in-mouse autograft model of human myeloma to study dormancy. The model relies on a transgenic enhanced green fluorescence protein (GFP) reporter to readily detect and accurately enumerate myeloma cells in host bone marrow by using fluorescence microscopy and flow cytometry as research tools. The model additionally relies on cytoplasmic labeling of myeloma cells with a lipophilic dye dubbed DiD that undergoes progressive dilution as cells divide and share label among daughter cells. Measurement of DiD fluorescence intensity enables distinction of dormant dye-retaining nonproliferating GFP+DiD+ cells and reactivated tumor-forming GFP+DiD− cells that lost the dye because of active cell cycling (see figure). An intermediate GFP+DiDLow cell type caught during transition from dormancy to reactivated growth can also be identified. Previous work from the investigator team demonstrated that myeloma dormancy is a reversible, dynamic state that is induced by cell-to-cell contact with osteoblasts in the endosteal bone marrow niche and released by osteoclast-dependent remodeling of the niche. Consistent with that, GFP+DiD+ dormant cells, when isolated from donor and reinjected in recipient mice, retain the ability to form new tumors and reenter dormancy.5
The nifty experimental model system outlined here is the foundation for the reported advance. Khoo et al use single-cell RNA sequencing to determine the global gene expression profile (transcriptome) of dormant 5TGM1 myeloma cells harvested from hematopoietic bone marrow samples of tumor-bearing mice. Data analysis revealed a novel transcriptional signature enriched with, unexpectedly, numerous upregulated genes that are normally involved in pathways of myeloid differentiation. This suggests that, in analogy to the epithelial-mesenchymal transition of solid cancers, dormant myeloma undergoes a lymphoid-myeloid transition of sorts. Of note, the interrogation of 2 independent clinical datasets demonstrated the significance of the dormancy-associated transcriptional signature for myeloma. For example, the discovery that individuals with the myeloma precursors condition, monoclonal gammopathy of undetermined significance, exhibited higher levels of the myeloid signature than patients with frank myeloma implicates immune and myeloid pathway genes in disease progression. Similarly, the twofold increase in overall survival of patients with myeloma that express the signature, relative to patients that do not, supports the contention that enhanced displacement of myeloma cells from osteoblastic dormancy niches promotes tumor growth and negatively affects outcome.
To evaluate whether dormancy-associated myeloid signature genes may lend themselves to the design and testing of molecularly targeted therapies aimed at eradicating “myeloma sleeper agents” and preventing disease relapse, Khoo et al focused on the most highly expressed gene, Axl, which encodes a synonymous receptor tyrosine kinase that is activated by growth arrest specific 6 expressed on osteoblasts. The authors showed that treatment of myeloma-bearing mice with broad-spectrum small-compound AXL inhibitors, such as cabozantinib and BMS-777607, releases 5TGM1 cells from dormancy, thus providing proof-of-concept for clinical studies that may determine whether the recently US Food and Drug Administration–approved receptor tyrosine kinase inhibitor, gilteritinib (50% inhibition for AXL is 0.73 nmol/L),6 might be more effective in patients with myeloma than cabozantinib, demonstrating no single-agent activity in a recent exploratory trial.7 As with all good preclinical studies addressing important knowledge gaps, the paper by Khoo et al raises the question whether the findings can be translated to human disease. A great deal of optimism in this regard is generated by independent earlier work in Orlowski’s group, which demonstrated that the osteoblastic niche supports quiescent human myeloma cells exhibiting enhanced tumor-forming potency and increased cancer stemness.8 Khoo et al should be commended for developing a cutting-edge technology to analyze dormant myeloma cells and paving the way for the molecular targeting of the dormancy-supporting bone marrow niche.
Conflict-of-interest disclosure: The authors declare no competing financial interests.