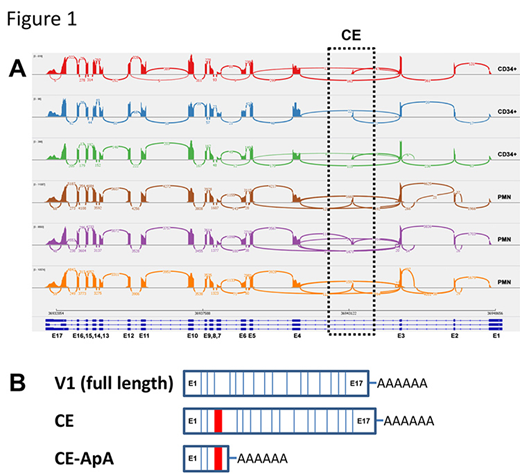
Increasing evidence suggests that alternative splicing of CSF3R occurs during normal granulopoiesis and is altered in certain pathologic conditions. To further examine CSF3R splicing, Sashimi plots from RNA-seq data were generated from CD34+ cells and neutrophils isolated from healthy donors (Figure 1A). The majority of splicing events were found to follow a pattern of enforced exon order resulting in production of CSF3R-V1, and to a lesser extent production of variants 3 and 4 that arise from alternative splicing of distal exonic sequences. However, a significant number of splicing events also mapped to a region within intron 3 (Figure 1A, black hashed box), suggesting a putative cryptic exon (CE).
RT-PCR was performed to amplify the region containing the putative CE and Sanger sequencing of the amplification products confirmed the presence of a 73 base pair CE between exons 3 and 4 (E3 and E4). The CE adds an in-frame stop codon that is far removed from the terminal exon; transcripts containing the CE would be predicted to be degraded by nonsense-mediated decay (NMD). We detected transcripts containing the CE sequence in HL-60 cells as well as in CD34+ cells, neutrophils, and monocytes from healthy donors. Cycloheximide had no appreciable effect on the CE-containing transcript, indicating this transcript does not undergo NMD.
One mechanism by which the CSF3R CE-containing transcripts could evade NMD would be through alternative polyadenylation (ApA). ApA commonly occurs in the 3'UTRs of mRNAs and is a mechanism for regulating differential transcript expression. ApA can also occur in introns and lead to production of either non-coding RNA or transcripts that produce truncated proteins. Using RT-PCR with a forward primer positioned across the E3:CE junction and a reverse primer positioned within the intronic sequence following the CE and Sanger sequencing of the amplified product, we detected the ApA transcript. The presence of the ApA transcript was confirmed by custom TaqMan assays capable of identifying each of the two predicted CE-containing CSF3R transcripts (Figure 1B). Transcription of either of the CE-containing transcripts would generate mRNAs with a very short open reading frame, and, hence, a protein of only 28 amino acids that is predicted to be signaling-incompetent. Production of the CE-containing CSF3R transcripts could quantitatively alter the amount of full-length CSF3R (V1) transcripts available for production of CSF3R protein, and therefore alter cellular responses to G-CSF.
We next examined cells from patients with AML to determine whether expression of the signaling-incompetent CE-containing CSF3R transcripts was altered. Real-time RT-PCR was performed on total RNA isolated from CD34+ cells from healthy donors and from patients with acute myeloid leukemia (AML) to quantitate "productive" CSF3R transcripts (signaling-competent CSF3R-V1 transcripts arising from correct splicing of E3 to E4) versus "non-productive" CSF3R transcripts (signaling-incompetent CSF3R+CE and CSF3R+CEApA transcripts) arising from inclusion of the CE. Significant changes in the relative amounts of CSF3R-V1, CSF3R+CE, and CSF3R+CEApA, and in overall CSF3R transcription were detected in AML samples compared to normal samples. We are currently investigating the effects of mutations in different splicing genes on the output of "productive" versus "non-productive" CSF3R transcripts.
In conclusion, we have identified two additional CSF3R transcripts generated by a CE following exon 3 that are expressed in human CD34+ cells and myeloid cells (Figure 1B). Notably, the expression of both CE-containing transcripts effectively decreases expression of the mature CSF3R mRNA, and by extension, of mature full-length CSF3R protein. Our data support a previously unrecognized mechanism of post-transcriptional regulation of CSF3R expression involving a CE and alternative splicing. Recently, widespread intronic polyadenylation of multiple genes including tumor suppressor genes was detected in cells from patients with CLL that resulted in expression of truncated mRNAs and proteins that functionally mimic somatic mutations, which was proposed as a novel mechanism for tumorigenesis. We suggest a similar mechanism involving CSF3R may play a role in AML.
Avalos:Best Practice-Br Med J: Patents & Royalties: receives royalties from a coauthored article on evaluation of neutropenia; Juno: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.