Introduction:
Current diagnostic standards for lymphoproliferative disorders include detection of clonal immunoglobulin (IG) and/or T cell receptor (TR) rearrangements, translocations, copy number alterations (CNA) and somatic mutations. These analyses frequently require a series of separate tests such as clonality PCR, fluorescence in situ hybridisation and/or immunohistochemistry, MLPA or SNParrays and sequencing. The EuroClonality-NGS DNA capture (EuroClonality-NDC) panel, developed by the EuroClonality-NGS Working Group, was designed to characterise all these alterations by capturing variable, diversity and joining IG and TR genes along with additional clinically relevant genes for CNA and mutation analysis.
Methods:
Well characterised B and T cell lines (n=14) representing a diverse repertoire of IG/TR rearrangements were used as a proficiency assessment to ensure 7 testing EuroClonality centres achieved optimal sequencing performance using the EuroClonality-NDC optimised and standardised protocol. A set of 56 IG/TR rearrangements across the 14 cell lines were compiled based on detection by Sanger, amplicon-NGS and capture-NGS sequencing technologies. For clinical validation of the NGS panel, clinical samples representing both B and T cell malignancies (n=280), with ≥ 5% tumour infiltration were collected from 10 European laboratories, with 88 (31%) being formalin fixed paraffin-embedded samples. Samples were distributed to the 7 centres for library preparation, hybridisation with the EuroClonality-NDC panel and sequencing on a NextSeq 500, using the EuroClonality-NDC standard protocol. Sequencing data were analysed using a customised version of ARResT/Interrogate, with independent review of the results by 2 centres. All cases exhibiting discordance between the benchmark and capture NGS results were submitted to an internal review committee comprising members of all participating centres.
Results:
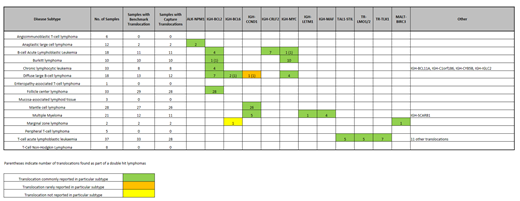
All 7 testing centres detected all 56 rearrangements of the proficiency assessment and continued through to the validation phase. A total of 10/280 (3.5%) samples were removed from the validation analysis due to NGS failures (n=1), tumour infiltration < 5% (n=7), and sample misidentification (n=2). The EuroClonality-NDC panel detected B cell clonality (i.e. detection of at least one clonal rearrangement at IGH, IGK or IGL loci) in 189/197 (96%) B cell malignancies. Seven of the 8 discordant cases were post-germinal centre malignancies exhibiting Ig somatic hypermutation. The EuroClonality-NDC panel detected T cell clonality (i.e. detection of at least one clonal rearrangement at TRA, TRB, TRD or TRG loci) in 70/73 (96%) T cell malignancies. In all 3 discordant cases analysis of benchmark PCR data was not able to detect clonality at any TR loci. Next, we examined whether the EuroClonality-NDC panel could detect clonality at each of the individual loci, resulting in sensitivity values of 95% or higher for all IG/TR loci, with the exception of those where limited benchmark data were available, i.e. IGL (n=3) and TRA (n=7). The specificity of the panel was assessed on benign reactive lesions (n=21) that did not contain clonal IG/TR rearrangements based on BIOMED-2/EuroClonality PCR results; no clonality was observed by EuroClonality-NDC in any of the 21 cases. Limit of detection (LOD) assessment to detect IG/TR rearrangements was performed using cell line blends with each of the 7 centres receiving blended cell lines diluted to 10%, 5.0%, 2.5% and 1.25%. Across all 7 centres the overall detection rate was 100%, 94.1%, 76.5% and 32.4% respectively, giving an overall LOD of 5%. Sufficient data were available in 239 samples for the analysis of translocations. The correct translocation was detected in 137 out of 145 cases, resulting in a sensitivity of 95%. Table 1 shows how translocations identified by the EuroClonality-NDC protocol were restricted to disease subtypes known to harbour those types of translocations. Analysis of CNA and somatic mutations in all samples is underway and will be presented at the meeting.
Conclusions:
The EuroClonality-NDC panel, with an optimised laboratory protocol and bioinformatics pipeline, detects IG and TR rearrangements and translocations with high sensitivity and specificity with a LOD ≤ 5% and provides a single end-to-end workflow for the simultaneous detection of IG/TR rearrangements, translocations, CNA and sequence variants.
Stamatopoulos:Janssen: Honoraria, Research Funding; Abbvie: Honoraria, Research Funding. Klapper:Roche, Takeda, Amgen, Regeneron: Honoraria, Research Funding. Ferrero:Gilead: Speakers Bureau; Janssen: Consultancy, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; EUSA Pharma: Membership on an entity's Board of Directors or advisory committees; Servier: Speakers Bureau. van den Brand:Gilead: Speakers Bureau. Groenen:Gilead: Speakers Bureau. Brüggemann:Incyte: Membership on an entity's Board of Directors or advisory committees; Amgen: Membership on an entity's Board of Directors or advisory committees; Roche: Consultancy. Langerak:Gilead: Research Funding, Speakers Bureau; F. Hoffmann-La Roche Ltd: Research Funding; Genentech, Inc.: Research Funding; Janssen: Speakers Bureau. Gonzalez:Roche: Honoraria, Research Funding; AstraZeneca: Consultancy, Honoraria, Research Funding, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.