Abstract
Background:
FISH is the gold standard for genetic characterization and risk stratification of multiple myeloma (MM) at diagnosis. Retrospective studies have shown the potential of whole genome sequencing (WGS) to gain new insights into MM genetics. With respect to clinical application, we prospectively performed FISH and WGS in parallel in patients (pts) with newly diagnosed MM (NDMM) and relapsed/refractory MM (RRMM) to evaluate advantages and limitations.
Patients and methods:
From bone marrow (BM) samples of 280 MM pts CD138+ cells were enriched by MACS. In 100 pts (30 NDMM, 70 RRMM) FISH (del(1p32), del(13q), del(17p), t(4;14), t(11;14), t(14;16), t(14;20), t(6;14), 1q gain, MYC rearrangement (MYCr) and trisomies 3, 5, 9) and WGS were performed. WGS: 150bp paired-end sequences were generated on NovaSeq 6000 (Illumina); median coverage 104x. Single nucleotide variants (SNV) were called with Strelka software, copy number variations (CNV) with GATK4 and structural variants (SV) with Manta. Technical artefacts and germline calls were reduced by a tumor/unmatched normal workflow with genomic DNA from a mix of anonymous donors. Variants with global population frequency >0.5% (gnomAD) were excluded.
Results:
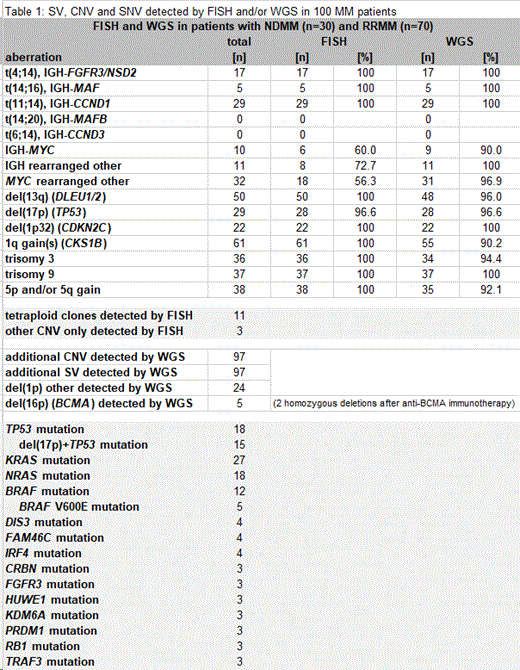
In 100/280 (36%) MM pts WGS could be performed in parallel to FISH in this cohort with a low median BM plasma cell infiltration (PCI) of 6% (range 1-95%). Likelihood of DNA yields sufficient for WGS increased with higher PCI (23% (1-95%) vs 3% (1-50%) in 100 pts with WGS vs 180 pts without sufficient DNA).
Comparison of FISH and WGS results for 100 pts showed 100% concordance for recurrent SV (table 1): t(11;14) in 29%, t(4;14) in 17% and t(14;16) in 5%. No pts harbored a t(14;20) but in one case FISH indicated another alteration of MAFB and WGS revealed MAFB rearranged to FAM46C resulting in an elevated MAFB expression.
Due to heterogeneous breakpoints FISH is less reliable for the detection of MYCr than WGS (24/42 (57%) detected by FISH vs 40/42 (95%) by WGS). Partners of MYC included IGH (n=10), FAM46C (n=4), IGK (n=3), IGL (n=3), FOXO3 (n=2) and IRF4 (n=1). Rare IGH rearrangements with partner genes such as IRF1, IRF4, ZBTB38 and ZFP36L1 were observed in 11 pts all identified by WGS, but only 8 (73%) were detected by FISH.
Total frequencies of CNV were del(13q) in 50%, del(17p) in 29%, del(1p32) in 22%, 1q gains in 61% and hyperdiploidy in 36%. WGS detected 95% of CNV (details table 1). CNV that were missed by WGS included one del(17p) and were all identified in small subclones with a median clone size of 10% (6-25%) in FISH.
Tetraploid MM, potentially harboring adverse prognostic implication, was identified in 11 pts by FISH but not in WGS.
However, in 97 pts WGS depicted additional SV and CNV compared to FISH. This includes aberrations of prognostic significance such as a TP53 deletion that was located adjacent to a 451kb gain on 17p. The FISH probe encompassing TP53 confirmed the 17p gain but masked the TP53 deletion. Also 1p deletions, which have been described to be a negative prognostic factor, were found more frequently by WGS with 24 pts harboring a del(1p) other than del(1p32) (locus evaluated by FISH) which was detected in 22 pts by both methods. Furthermore, as WGS cannot only detect SV and CNV but also SNV, WGS allows screening for potentially clinically exploitable drug targets, e.g. BRAF V600E mutation (5 pts in our cohort) or resistance associated mutations such as in cereblon for immunomodulatory compounds (CRBN mutation in 3 pts) as well as biallelic events in TP53 (del(17p)+TP53 mutation), detected in 15 of our pts suggesting worst prognosis for affected pts. WGS in RRMM can shed light on the genetic background of relapses. Of note, homozygous del(16p) inducing loss of the treatment target were first described using our dataset in two patients with acquired resistance toward anti-BCMA CAR T cells or bispecific antibody therapy.
Conclusions:
FISH is the preferred approach in pts with BM PCI <10% and for the detection of clones <15% or variant ploidy levels. WGS is superior to FISH regarding the identification of biallelic events and rearrangements with heterogeneous breakpoints and rare partners. But even more, WGS can address various questions in follow up in terms of increase of complexity, target specific therapy, therapy failure and relapse. Thus, WGS allows a holistic insight into the genetics of MM and provides crucial information for clinical decision making.
Kern: MLL Munich Leukemia Laboratory: Other: Part ownership. Haferlach: MLL Munich Leukemia Laboratory: Other: Part ownership. Einsele: Janssen, Celgene/BMS, Amgen, GSK, Sanofi: Consultancy, Honoraria, Research Funding. Haferlach: MLL Munich Leukemia Laboratory: Other: Part ownership.