Much of our knowledge of hematopoiesis in health and disease over the last 50 years has been the product of early adoption of technological breakthroughs, at least in part as a result of the relative ease of accessing patient material compared with many other clinical specialties. This early adoption of technology included methods for the examination of phenotype and function of single cells, alongside detection of chromosomal rearrangements in single malignant hematopoietic cells. Fluorescence-activated cell sorting (FACS) using monoclonal antibodies1,2 allowed isolation and characterization of cells defined by specific combinations of surface markers, while in vitro colony assays3,4 and single-cell transplantation permitted functional assessment of lineage and proliferative potential.5-7 The advent of single-cell reverse-transcription polymerase chain reaction8 extended our understanding of the molecular mechanisms underpinning hematopoietic cell choice. By leveraging the resulting granular knowledge of single-cell function and potential, hematology research is now reaping the rewards of early adoption of new single-cell genomics techniques.
The study of normal hematopoiesis has been transformed in recent years by the application of single-cell genomics technologies that permit unbiased interrogation of molecular profiles at unprecedented resolution. Using massively parallel single-cell RNA (scRNA) sequencing combined with indexed FACS sorting, chromatin profiling, and functional assays, Paul et al9 were able to characterize the transcriptional heterogeneity and differentiation potential of myeloid progenitors. An unbiased computational approach uncovered transient intermediate fates within the myeloid progenitor compartment demonstrating the utility of scRNA sequencing in systematically profiling plastic and dynamic cell populations. Microfluidics-based droplet encapsulation techniques have expanded the throughput of single-cell technologies, now permitting the analysis of >104 cells per experiment.10 Computational analysis of thousands of transcriptomes identifies rare populations that would be lost in bulk assays, while enabling a tissue-level overview.11,12 Arranging single-cell profiles into coherent landscapes progressing from multipotent through unilineage progenitors to mature populations facilitates identification of putative decision points at which cells embark on a specific differentiation potential.13,14 These landscapes form part of larger reference atlases that map developing hematopoiesis across the life span of the organism15,16 and provide comparator datasets for the disease state.17
We provide a commentary to 4 review articles published in this issue of Blood exploring the status and future potential of single-cell genomics for studying hematological malignancy.
Asiri Ediriwickrema, Andrew J. Gentles, and Ravindra Majeti, “Single-cell genomics in AML: extending the frontiers of AML research”
Ilaria Iacobucci, Matthew T. Witkowski, and Charles G. Mullighan, “Single-cell analysis of acute lymphoblastic and lineage-ambiguous leukemia: approaches and molecular insights”
Adi Nagler and Catherine J. Wu, “The end of the beginning: application of single-cell sequencing to chronic lymphocytic leukemia”
Jennifer Mary O’Sullivan, Adam J. Mead, and Bethan Psaila, “Single-cell methods in myeloproliferative neoplasms: old questions, new technologies”
Ediriwickrema et al describe the contribution of single-cell genomics to addressing unanswered questions in the pathophysiology and management of acute myeloid leukemia (AML), emphasizing progress in unraveling the disease heterogeneity that may drive treatment resistance and relapse. Although the treatment landscape for AML has recently diversified after decades of frustratingly negative large-scale clinical trials,18 risk stratification based on bulk molecular profiling and the perennial problem of disease relapse limit the full potential of newly introduced agents. Referencing a number of seminal recent studies that have reconstructed the clonal architecture and cellular state of AML through treatment,19-21 the authors argue that AML research is on the threshold of a new frontier integrating mutational, transcriptional, epigenomic, and surface phenotype at unprecedented resolution to address the role of the leukemic stem cell, microenvironment, and selection pressure in disease initiation, treatment response, and progression, ultimately improving our ability to tailor therapy.
In their review on acute lymphoblastic leukemia, Iacobucci et al highlight the advantages of single-cell sequencing over bulk next-generation sequencing in advancing the molecular taxonomy of acute lymphoblastic leukemia (ALL) and correlating single-cell transcriptional signatures of ALL blasts with normal B-cell precursors to address the identification of the “cell of origin.” In ALL, unlike AML, immune therapy with bispecific antibody and chimeric antigen receptor T-cell therapy have revolutionized the treatment landscape in the relapse setting, and single-cell studies have shown the potential to identify new targetable pathways to potentiate the long-term antileukemic effect.
In their review on chronic lymphocytic leukemia (CLL), Nagler and Wu discuss studies describing the clonal evolution of the disease through a time-course encompassing sequential treatments to which most patients may be exposed during the protracted disease course. Taking advantage of epigenomic data and mitochondrial mutations allows construction of CLL lineage trees and tracking of subclonal populations in elegant work that permits characterizing nongenetic mechanisms of clonal evolution in disease progression, transformation, and therapy resistance.22,23 The dependence of CLL on immune cells in the microenvironment has long been recognized, and the authors comprehensively review single-cell studies focused on this cross talk and its interaction with treatment, making the case that the diverse microenvironments of this multi-sited disorder may benefit from novel spatial single-cell profiling techniques.
O’Sullivan et al review how the use of single-cell genomics in myeloproliferative neoplasms (MPNs) provides insight into the lineage bias introduced by malignant transformation of hematopoietic stem and progenitor cells (HSPCs) in these disorders, which is read out as increased abundance of mature blood lineages. Single-cell analysis of these biases, combined with targeted single-cell mutational analysis, identifies aberrant programs driving inflammation and fibrosis in myelofibrosis and identifies therapeutic vulnerabilities unique to mutant HSPCs, by comparison with wild-type internal control HSPCs, whose own transcriptional profiles can be influenced by the presence of disease through extrinsic mechanisms.24,25 Further, the authors describe the emerging potential of single-cell mutation detection and single-cell spatial profiling techniques to predict disease progression and conclude with a persuasive argument for prospective inclusion of single-cell technologies in future clinical trials, with a specific focus in MPN on identifying and eliminating the MPN stem cell.
How far are we from translating these techniques into clinical practice and where could the most value be added? In the short to medium term, as throughput approximates that of high-resolution flow cytometry, likely the greatest added value for single-cell genomics may be gained from clonal tracking and assessment of remission or residual disease, complementing current flow cytometry and bulk sequencing minimal residual disease methods by detecting rare populations, demonstrating subclonal hierarchies and nongenomic heterogeneity, and predicting prognosis or response to further treatment. Although eradication of minimal residual disease is of greatest prognostic significance in the acute leukemias, monitoring cell state alongside mutation status at progression or relapse is likely to carry prognostic and treatment value for all hematological malignancies. Indeed, in the chronic disease setting where disease eradication is rare or impossible, such that persistent disease is detectable at higher levels, the level of resolution of current single-cell technologies may be poised to offer the greatest value.
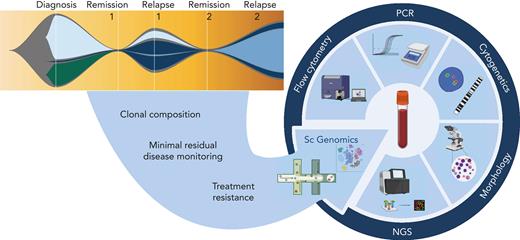
Common themes emerging from all 4 reviews in the current series are of tumor heterogeneity and microenvironmental factors at diagnosis that may predict treatment response and tumor evolution. In the medium to long term, further single-cell multiomic and spatial studies characterizing mechanisms of disease initiation and progression are likely to elucidate treatment vulnerabilities and novel risk stratifications that will inform treatment choice at diagnosis and relapse. At present, single-cell genomics techniques incur significant financial cost. Moreover, technical limitations necessitate an extensive analysis pipeline that is not yet standardized, thus generating additional cost in skilled data analysis hours and a lead time from sample acquisition to availability of clinically relevant individualized biomarkers. Further, generalizability is currently limited because of necessarily small cohort analyses that reveal significant inter- and intra-patient heterogeneity, rendering essential the public sharing of datasets to allow meta-analysis and complementary studies from bulk sequencing of larger datasets.26 To make the transition from research tool to standard-of-care testing, clinical trials are required adding complementary single-cell studies to current bulk methods of diagnosis and monitoring. Notwithstanding current limits, it is clear that huge gains will ensue from clinical integration of these techniques into personalized oncology (Figure 1), much in the same way as next-generation sequencing has transitioned from Nature front cover to influencing routine clinical practice in little over a decade.
Potential translational role of single-cell genomics in hemato-oncology diagnostics. Complementing the current repertoire of tests applied at diagnosis, remission, and relapse, we anticipate that the addition of single-cell genomics techniques has the potential to delineate clonal composition, track rare populations through remission and relapse, and identify transcriptional signatures that predict treatment response or resistance. NGS, next-generation sequencing; PCR, polymerase chain reaction. Figure created with BioRender.com.
Potential translational role of single-cell genomics in hemato-oncology diagnostics. Complementing the current repertoire of tests applied at diagnosis, remission, and relapse, we anticipate that the addition of single-cell genomics techniques has the potential to delineate clonal composition, track rare populations through remission and relapse, and identify transcriptional signatures that predict treatment response or resistance. NGS, next-generation sequencing; PCR, polymerase chain reaction. Figure created with BioRender.com.
Acknowledgments
The authors acknowledge research funding from Cancer Research UK, Blood Cancer UK, UK Research and Innovation Medical Research Council, and Wellcome.
Conflict-of-interest disclosure: The authors declare no competing financial interests.