Key Points
ID1 deficiency in the AML BMM disrupts the progression of AML, which is restored by Angptl7.
ID1 interacts with RNF4, attenuates SP1 ubiquitination, and contributes to the expression of ANGPTL7 in AML BM mesenchymal stem cells.
Abstract
The bone marrow microenvironment (BMM) can regulate leukemia stem cells (LSCs) via secreted factors. Increasing evidence suggests that dissecting the mechanisms by which the BMM maintains LSCs may lead to the development of effective therapies for the eradication of leukemia. Inhibitor of DNA binding 1 (ID1), a key transcriptional regulator in LSCs, previously identified by us, controls cytokine production in the BMM, but the role of ID1 in acute myeloid leukemia (AML) BMM remains obscure. Here, we report that ID1 is highly expressed in the BMM of patients with AML, especially in BM mesenchymal stem cells, and that the high expression of ID1 in the AML BMM is induced by BMP6, secreted from AML cells. Knocking out ID1 in mesenchymal cells significantly suppresses the proliferation of cocultured AML cells. Loss of Id1 in the BMM results in impaired AML progression in AML mouse models. Mechanistically, we found that Id1 deficiency significantly reduces SP1 protein levels in mesenchymal cells cocultured with AML cells. Using ID1-interactome analysis, we found that ID1 interacts with RNF4, an E3 ubiquitin ligase, and causes a decrease in SP1 ubiquitination. Disrupting the ID1-RNF4 interaction via truncation in mesenchymal cells significantly reduces SP1 protein levels and delays AML cell proliferation. We identify that the target of Sp1, Angptl7, is the primary differentially expression protein factor in Id1-deficient BM supernatant fluid to regulate AML progression in mice. Our study highlights the critical role of ID1 in the AML BMM and aids the development of therapeutic strategies for AML.
Introduction
Acute myeloid leukemia (AML) is a genetically heterogeneous hematological malignant disease with a poor survival rate.1-3 Over the past few years, many discoveries of potential genetic and epigenetic lesions within AML cells have provided hope for clinical advancement.4-8 However, in addition to the cell-autonomous functions, non-cell-autonomous mechanisms are also crucial causes of AML.9-13 These observations suggest that elucidating the mechanisms by which the bone marrow microenvironment (BMM) maintains leukemia stem cells (LSCs) may provide potential therapeutic strategies for AML.
Inhibitor of DNA binding 1 (ID1), a member of helix-loop-helix proteins, lacks a DNA-binding motif.14,15 Although the helix-loop-helix domain of the ID1 protein is involved in the majority of its interaction events, additional motifs located in its N- and C-terminal regions are required for the identification of varying partners. We previously uncovered that ID1, a target gene for AML1-ETO,16 interacts directly with AKT1 via its C-terminal region to promote AKT1 phosphorylation.17 It has been reported that Smurf2, an E3 ubiquitin ligase, mediates ubiquitination of Id1 and that the deletion of the N terminal of Id1 abolishes the interaction between Id1 and Smurf2.18,19 Moreover, the deubiquitinating enzyme USP1 can associate with, and deubiquitinate, ID1 in mesenchymal stem cells (MSCs), thus preserving their stem cell program in osteosarcoma.20
Id1 is a key regulator of hematopoietic stem cell (HSC) behavior, because Id1 deletion reduces the self-renewal capacity of HSCs and increases their propensity for myeloid differentiation.21,22 Elevated levels of ID1 messenger RNA (mRNA) and protein have been found in many tumor types, in which they have often been associated with poor prognosis.23-26 We previously identified that Id1 controls t(8;21) leukemia initiation and progression.17 Moreover, our previous studies have shown that fetal liver and BM hematopoietic stem and progenitor cells (HSPCs) show different dependence on Id1 for MLL-AF9–mediated leukemogenesis. Id1 ablation in fetal liver HSPCs significantly attenuates the development of leukemia, whereas Id1 loss in BM HSPCs accelerates leukemogenesis. These investigations demonstrated a distinct effect of Id1 on the initiation and development of leukemia between infant and postnatal MLL-AF9–driven leukemia.27,28
In addition to intrinsically enhancing HSC/LSC proliferation, ID1 is widely expressed in BMM cells and affects hematopoiesis.29-32 BMPs, the transforming growth factor–β family members, have vital roles in directing cell fate decision of mesenchymal cells33,34 and elicit their cellular effects by activating ID1 expression.35,36 Previous studies have shown that Id1 expression in the BMM is critical for normal hematopoietic development.37 Recent studies have shown that Id1 is required for the survival and steady-state regeneration of endothelial cells, which sustain HSC quiescence and survival.38
Based on these reports, we hypothesize that Id1 may also regulate AML development via nonautonomous cell mechanisms. We demonstrate that an Id1-deficient BMM exhibit impaired AML progression. We observed significant cell cycle arrest in AML cells cocultured with ID1−/− mesenchymal cells compared with wild-type mesenchymal cells. The highly expressed ID1 in mesenchymal cells is induced by BMP6 in the coculture system. The ID1-RNF4 interaction weakens SP1 ubiquitination and subsequently activates the transcription of ANGPTL7 in mesenchymal cells. Moreover, intra-BM transfusion of Angptl7 significantly promotes the development of AML in an Id1-deficient BMM in vivo. In summary, we identify a novel nonautonomous cell mechanism by which ID1 expressed in AML BMM sustains LSCs, with possible therapeutic implications for AML.
Materials and methods
Coculture assay
An indirect coculture system was used to investigate the interaction between the human-derived BM mesenchymal stem cell (BMSC) cell line HS-5 and leukemia cells and the functional changes in BMSCs and leukemia cells. For the coculture assay, 1 × 105 HS-5 cells were preseeded in the lower compartment of a 6-well plate for 24 hours, and 1 × 105 leukemia cells were cultured at the top of the transwell membrane. After 4 days, the leukemia cells and HS-5 cells were collected separately.
Results
ID1 is highly expressed in AML-MSCs, especially in AML subtypes M2 and M5
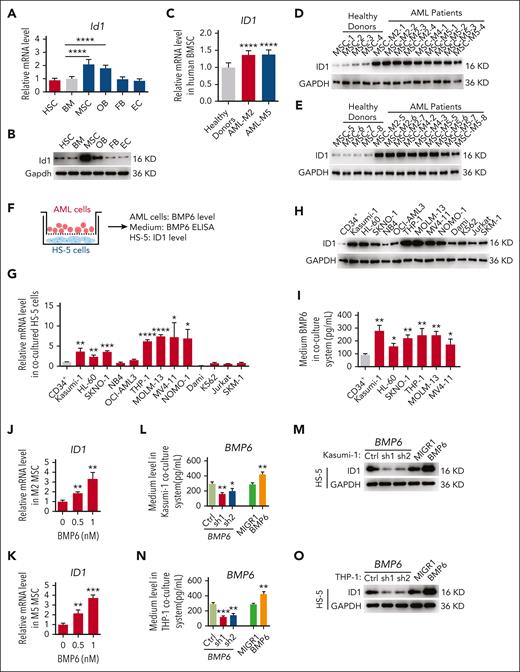
To determine the role of Id1 in the BMM, we first analyzed the expression patterns of Id1 in HSCs and different types of BMM cells via quantitative polymerase chain reaction (qPCR) and western blot assays. The results showed that the Id1 was expressed in HSCs and several BMM cells (MSC, osteoblast, fibroblast, and endothelial cells; Figure 1A-B; supplemental Figure 1A-D, available on the Blood website). Strikingly, MSCs had higher mRNA and protein levels of Id1. Because Id1 is highly expressed in BMM cells, it would be interesting to determine whether Id1 regulates leukemia progression through a nonautonomous cellular role. To this end, human-derived BMSCs from healthy donors and patients with AML (HD-MSCs and AML-MSCs, respectively) were collected and immunophenotyped via surface antigen expression flow cytometry (supplemental Figure 1E). We analyzed HD-MSCs and AML-MSCs using qPCR and western blot assays. The results showed that the ID1 mRNA and protein levels were higher in AML-MSCs than in HD-MSCs (Figure 1C-E). This observation leads us to question whether the MSCs of all AML subtypes have increased ID1 levels. To address this important question, we set up a coculture assay, in which HS-5 cells were cultured together with leukemia cells of different genetic makeups (Figure 1F). After 4 days, HS-5 cells were collected from the coculture system, and qPCR and western blot assays were performed. Interestingly, a significant elevated ID1 expression level in HS-5 cells cultured with M2-subtype AML cells (Kasumi-1, HL-60, and SKNO-1) and M5-subtype AML cells (THP-1, MOLM-13, MV4-11, and NOMO-1) compared with those cultured with CD34+ cells were observed (Figure 1G-H). We analyzed the AML data from The Cancer Genome Atlas Program database,39 which showed that patients with AML subtype M2 or M5 with high ID1 expression had a significantly worse overall survival and shorter median survival than patients with low ID1 expression (supplemental Figure 1F). We further investigated the ID1 expression level in AML-MSC using qPCR, and the results showed that ID1 mRNA levels were significantly downregulated in patients with AML in remission (supplemental Figure 1G).
To investigate the potential mechanism of ID1 upregulation in AML-MSCs, we analyzed the binding peaks of AML1-ETO15 and MLL-AF940 on several cytokine genes that functionally activate the expression of ID1.35,36,41 The results showed that AML1-ETO and MLL-AF9 proteins bind to the promoter or gene body region of BMP1, BMP6, BMP8A, and interleukin-1β (IL-1β) (supplemental Figure 2). We knocked down AML1-ETO in Kasumi-1 cells and observed a significant downregulation of BMP6 and IL-1β (supplemental Figure 3A-B). Consistent with this finding, Bmp6 and Il-1β expression is significantly upregulated in 32D cells transduced with retrovirus expressing MLL-AF9 (supplemental Figure 3C-D). Furthermore, we analyzed the expression of ID1 in patient-derived BMSCs supplemented with recombinant BMP6 or IL-1β and found a more significant ID1 upregulation after BMP6 treatment (supplemental Figure 3E-F).
We found that BMP6 was significantly upregulated in Kasumi-1, SKNO-1, THP-1, and MOLM-13 groups in the medium of the coculture system (Figure 1I). AML-MSCs derived from patients with AML subtype M2 or M5 were treated with recombinant BMP6. The qPCR analysis showed that recombinant BMP6 increased the expression of ID1 in BMSCs (Figure 1J-K). To determine whether ID1 in mesenchymal cells is regulated by BMP6 secreted from AML cells, we used short hairpin RNA (shRNA) against BMP6 to knock down BMP6 in Kasumi-1 and THP-1 cells and observed significant downregulation of ID1 protein levels in HS-5 cells cocultured with these cells. Consistent with these findings, the ID1 protein levels in HS-5 cells were significantly upregulated after cocultured with retrovirus-transduced Kasumi-1 and THP-1 cells overexpressing BMP6 (Figure 1L-O). To confirm these findings in patients with AML, matched BM cells and MSCs derived from patients with AML were used (supplemental Figure 3G). We knocked down BMP6 in CD34+ leukemia blast cells derived from patients with AML subtypes M2, M4, and M5 and found that downregulation of BMP6 in CD34+ leukemia blast cells significantly reduced ID1 mRNA levels in matched AML-MSCs (supplemental Figure 3H-J). We further analyzed the expression of ACVR1, the receptor of BMP6,42-45 in HD-MSC and AML-MSC RNA-sequencing data,46 and found no significant difference in ACVR1 expression between HD-MSCs and AML-MSCs (supplemental Figure 3K). Consistent with this finding, by using qPCR, we observed no significant difference in ACVR1 expression in M2/M5 AML-MSCs compared with that in HD-MSC controls (supplemental Figure 3L). Together, these results suggest that elevated BMP6 levels in the M2/M5 subtype AML cells induce the expression of ID1 in MSCs. We narrowed our investigation to focus on AML models driven by AML1-ETO/MLL-AF9.
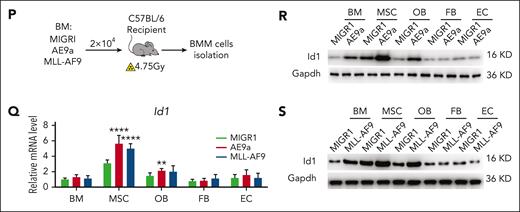
To further validate the possible role of ID1 in these 2 subtypes of AML BMM, we performed in vivo secondary transplantation assays. We transplanted 2 × 104 AML1-ETO9a (AE9a)- or MLL-AF9–expressing leukemia cells into sublethally irradiated recipient mice (Figure 1P). Two weeks after transplantation, we analyzed the BMM cells of mice with AML and control mice, using qPCR and western blot assays. The results showed that the Id1 mRNA and protein levels were higher in the AML BMM, especially in MSCs (Figure 1Q-S). Taken together, these results suggest a potential nonautonomous cellular role for ID1 in AML1-ETO/MLL-AF9–driven AML.
ID1 in BMSCs promotes AML cell proliferation
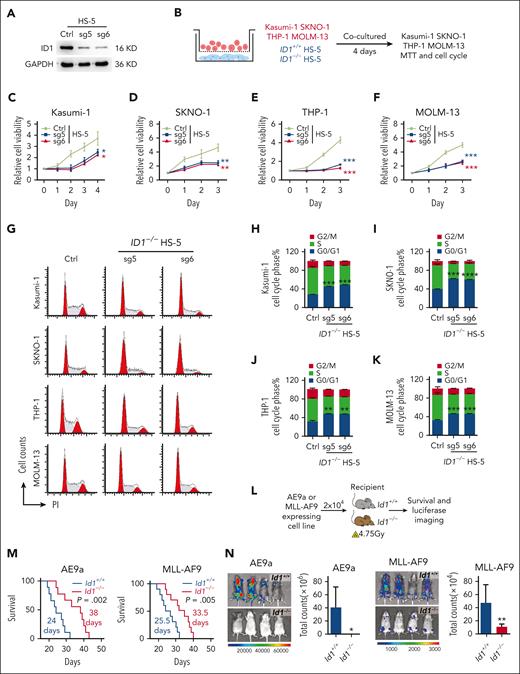
The BMSC cell lines HS-5, MS-5, and OP9 now constitute the key stromal cell lines used to promote HSPC expansion or differentiation in vitro.47-56 Here, we further investigated the nonautonomous cellular role of ID1 in AML, using a model of an in vitro coculture system. We targeted ID1 with single guide RNA and shRNA, which exhibit significant silencing of ID1 at the protein level in HS-5, MS-5, and OP9 cells (Figure 2A; supplemental Figure 4A). We performed coculture of Kasumi-1, SKNO-1, THP-1, and MOLM-13 cells with HS-5 cells for 4 days before 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTT) assays (Figure 2B). The results indicated that deletion of ID1 in HS-5 cells significantly suppressed the proliferation of AML cells nonautonomously (Figure 2C-F). In addition, the flow cytometry analysis showed that knocking out ID1 in HS-5 cells induced cell cycle arrest at G0/G1 phase in Kasumi-1, SKNO-1, THP-1, and MOLM-13 cells (Figure 2G-K). Similarly, Id1 deficiency in MS-5 or OP9 cells induced cell cycle arrest at G0/G1 phase in AE9a- and MLL-AF9–expressing murine AML cells (supplemental Figure 4B-C). Next, we sought to evaluate the nonautonomous cellular role of Id1 in AML progression in sublethally irradiated recipient mice (Figure 2L). The Id1−/− mice receiving AE9a (n = 9) or MLL-AF9 (n = 10) cell line had a longer life span than the Id1+/+ group (Figure 2M). Because these cells contain luciferase, we detected more intense bioluminescent signals in the Id1+/+ mice compared with the Id1−/− recipients after the transplantation (Figure 2N). Together, Id1 knockout in BMSCs primarily inhibits the growth of AML cell lines and induces cell cycle arrest in these cells.
ID1 knockout in BMSCs suppresses AML cell proliferation in vitro and in vivo. (A)The western blot analysis of ID1 expression in HS-5 cells with ID1 knockout and in control cells. (B-F) MTT assays show that ID1 knockout in HS-5 cells significantly inhibits the proliferation of cocultured Kasumi-1, SKNO-1, THP-1, and MOLM-13 cells (n = 3). (G-K) Cell cycle analysis on Kasumi-1, SKNO-1, THP-1, and MOLM-13 cells cocultured with ID1+/+ or ID1−/− HS-5 cells for 4 days (n = 3). (L) The strategy of AE9a- and MLL-AF9–expressing cell line transplantation. (M) Id1−/− recipients showed a significantly longer survival time than Id1+/+ recipients after undergoing transplantation with AE9a (n = 9; median survival, 24 vs 38 days) or MLL-AF9 cell line (n = 10; median survival, 25.5 vs 33.5 days). (N) The in vivo bioluminescence imaging and quantification analysis shows that the Id1−/− BMM impairs leukemia progression in recipients that received transplantation with AE9a or MLL-AF9 cell lines that are luciferase positive (n ≥ 4). ∗P < .05; ∗∗P < .01; ∗∗∗P < .005; ∗∗∗∗P < .001. Ctrl, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PI, propidium iodide; sg, single guide.
ID1 knockout in BMSCs suppresses AML cell proliferation in vitro and in vivo. (A)The western blot analysis of ID1 expression in HS-5 cells with ID1 knockout and in control cells. (B-F) MTT assays show that ID1 knockout in HS-5 cells significantly inhibits the proliferation of cocultured Kasumi-1, SKNO-1, THP-1, and MOLM-13 cells (n = 3). (G-K) Cell cycle analysis on Kasumi-1, SKNO-1, THP-1, and MOLM-13 cells cocultured with ID1+/+ or ID1−/− HS-5 cells for 4 days (n = 3). (L) The strategy of AE9a- and MLL-AF9–expressing cell line transplantation. (M) Id1−/− recipients showed a significantly longer survival time than Id1+/+ recipients after undergoing transplantation with AE9a (n = 9; median survival, 24 vs 38 days) or MLL-AF9 cell line (n = 10; median survival, 25.5 vs 33.5 days). (N) The in vivo bioluminescence imaging and quantification analysis shows that the Id1−/− BMM impairs leukemia progression in recipients that received transplantation with AE9a or MLL-AF9 cell lines that are luciferase positive (n ≥ 4). ∗P < .05; ∗∗P < .01; ∗∗∗P < .005; ∗∗∗∗P < .001. Ctrl, control; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PI, propidium iodide; sg, single guide.
Id1 expression in host BMM results in accelerated AML in vivo
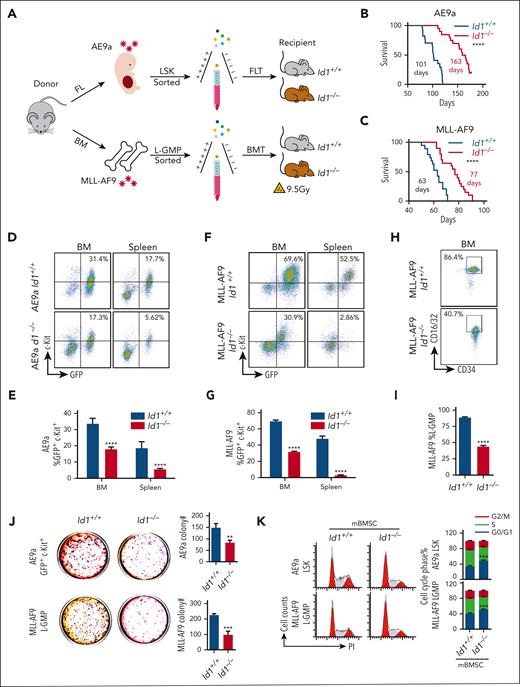
LSCs, characterized by their unlimited self-renewal/repopulation potential, are the root cause of disease initiation and progression as well as treatment failure and relapse in AML.57,58 To assess whether Id1 can regulate LSCs in a non-cell-autonomous manner via the BMM, we used AE9a/MLL-AF9–driven AML mouse transplantation models. We isolated E14.5 fetal liver cells and adult BM cells from donor mice transduced with retrovirus expressing AE9a or MLL-AF9, respectively. After the sorting of AE9a-expressing lineage (Lin)− Sca-1+ c-Kit+ cells (LSKs) and MLL-AF9–expressing leukemia–granulocyte macrophage progenitor (L-GMP) cells, the lethally irradiated Id1+/+ and Id1−/− recipient mice were injected with these cells via the tail vein (Figure 3A). In the mouse model of AE9a- or MLL-AF9–driven leukemia, the Id1−/− recipients showed a significant longer median survival than the Id1+/+ recipient mice (n ≥ 17; Figure 3B-C). Four weeks after transplantation, the Id1−/− recipients had significant lower white blood cell counts and smaller spleen size than Id1+/+ recipient mice (supplemental Figure 5A-E). The flow cytometry analysis showed that BM cells of Id1−/− recipient mice contained far fewer green fluorescent protein–positive (GFP+) c-Kit+ leukemia blast cells (Figure 3D-G), and Id1−/− recipients with MLL-AF9–driven leukemia significantly decreased the percentage of L-GMP cells (Figure 3H-I). Furthermore, the BM and spleen of the Id1−/− recipients with AE9a-driven leukemia contained far fewer GFP+Mac-1− cells (supplemental Figure 5F-G). The frequencies of GFP+Mac-1+ cells in the BM and spleen of Id1−/− recipients with MLL-AF9–driven leukemia were lower than those in the BM and spleen of Id1+/+ recipients (supplemental Figure 5H-I). Indeed, Wright and hematoxylin and eosin staining assays revealed that many leukemia blasts emerged in the BM and spleen of Id1+/+ recipients (supplemental Figure 5J-M). There is no significant effect on homing ability of LSCs between Id1+/+ and Id1−/− recipients (supplemental Figure 5N-P).
Id1 expression in the host BMM leads to AML acceleration. (A) The strategy of FLT or BMT. (B) Survival curves of Id1+/+ and Id1−/− recipients that underwent transplantation with AE9a-expressing fetal liver LSKs primarily (n ≥ 17; median survival, 101 vs 163 days). (C) Survival curves of Id1+/+ and Id1−/− recipients that underwent transplantation with MLL-AF9–expressing BM L-GMP cells primarily (n ≥ 19; median survival, 63 vs 77 days). (D-G) Representative flow cytometry profiles and quantification of the frequencies of the GFP+c-Kit+ leukemia blast cells in the BM and spleen of the indicated mice at 4 weeks after FLT or BMT (n = 10). (H-I) Representative flow cytometry profiles and quantification of the frequencies of the L-GMP cells in the BM of the indicated mice at 4 weeks after BMT (n = 10). (J) Colony-forming unit (CFU) assays analyzing the GFP+c-Kit+ leukemia blast cells sorted from FLT recipients BM or L-GMP cells sorted from BMT recipients BM. Average number of colonies generated from 1000 cells (n = 3). (K) Cell cycle analysis on AE9a-expressing LSKs and MLL-AF9–expressing L-GMP cells cocultured with Id1+/+ BMSCs or Id1−/− BMSCs for 5 days (n = 3). ∗P < .05; ∗∗P < .01; ∗∗∗P < .005; ∗∗∗∗P < .001.
Id1 expression in the host BMM leads to AML acceleration. (A) The strategy of FLT or BMT. (B) Survival curves of Id1+/+ and Id1−/− recipients that underwent transplantation with AE9a-expressing fetal liver LSKs primarily (n ≥ 17; median survival, 101 vs 163 days). (C) Survival curves of Id1+/+ and Id1−/− recipients that underwent transplantation with MLL-AF9–expressing BM L-GMP cells primarily (n ≥ 19; median survival, 63 vs 77 days). (D-G) Representative flow cytometry profiles and quantification of the frequencies of the GFP+c-Kit+ leukemia blast cells in the BM and spleen of the indicated mice at 4 weeks after FLT or BMT (n = 10). (H-I) Representative flow cytometry profiles and quantification of the frequencies of the L-GMP cells in the BM of the indicated mice at 4 weeks after BMT (n = 10). (J) Colony-forming unit (CFU) assays analyzing the GFP+c-Kit+ leukemia blast cells sorted from FLT recipients BM or L-GMP cells sorted from BMT recipients BM. Average number of colonies generated from 1000 cells (n = 3). (K) Cell cycle analysis on AE9a-expressing LSKs and MLL-AF9–expressing L-GMP cells cocultured with Id1+/+ BMSCs or Id1−/− BMSCs for 5 days (n = 3). ∗P < .05; ∗∗P < .01; ∗∗∗P < .005; ∗∗∗∗P < .001.
GFP+c-Kit+ cells expressing AE9a and L-GMP cells expressing MLL-AF9 were sorted from the BM of Id1+/+ and Id1−/− recipients with AML, and colony formation assays were performed to assess the self-renewal capacity of these cells. Strikingly, Id1 deficiency in the BMM significantly decreased the size and number of colonies (Figure 3J; supplemental Figure 5Q-R). To investigate whether Id1 in BMSCs influences AML progression, we cocultured AE9a-expressing LSKs or MLL-AF9–expressing L-GMP cells with Id1+/+ and Id1−/− primary BMSCs for 4 days in vitro. Cell cycle analysis indicates that Id1 deletion in BMSCs remarkably induces G0/G1 phase arrest of AE9a-expressing LSKs or MLL-AF9–expressing L-GMP cells (Figure 3K). These results suggest that Id1 in BMSCs promotes LSCs self-renewal in a nonautonomous cellular role.
Because loss of Id1 has a differential effect between fetal liver– and adult BM–derived leukemias,27,28 it is important to determine how Id1−/− fetal liver or Id1−/− BM cells are altered if injected into Id1 deficient mice. To address this important issue, we performed MLL-AF9 fetal liver transplantation and BM transplantation. We isolated Id1+/+ and Id1−/− E14.5 fetal liver cells or BM cells from Id1+/+ and Id1−/− donors and these cells were transduced with retrovirus expressing MLL-AF9. The Id1+/+ and Id1−/− MLL-AF9 expressing GFP+ L-GMP cells were injected into the tail vein of lethally irradiated Id1+/+ or Id1−/− recipient mice (supplemental Figure 6A). In fetal liver transplantation, which resembles MLL-AF9 infant leukemia, there was an additive effect on delayed AML progression and a reduction in the percentage of L-GMP cells when Id1−/− fetal liver L-GMP cells were transplanted into Id1−/− recipients. In BM transplantation, which resembles MLL-AF9 adult leukemia, there is an additive effect on delayed AML progression and a decrease in the percentage of L-GMP cells when Id1+/+ BM L-GMP cells are transplanted into Id1−/− recipients; this is consistent with what we previously uncovered that Id1 loss in BM HSPCs accelerates leukemogenesis (supplemental Figure 6B-C).
ANGPTL7 stimulates AML cell expansion in vitro
To investigate protein changes in BM supernatant fluid (BMSF) of Id1+/+ and Id1−/− recipients with AML, we applied tandem mass tag–based proteomics (Figure 4A). In total, 6973 proteins were quantified in each single sample, of which 371 differentially expressed proteins were identified (supplemental Figure 7A-B). The top 20 downregulated and upregulated secreted proteins in BMSF proteomics are shown in a heatmap (Figure 4B). The upregulated factors (Mup2, Tmem106b, Fxyd1, and Cd79a) in Id1−/− BMSF were validated via function analysis and qPCR analysis (supplemental Figure 7C). Because these factors are present at very low levels in the extracellular layer and their role in hematopoiesis and leukemogenesis has rarely been reported, we did not select them as targets for study. For the downregulated factors in Id1−/− BMSF, we validated the expression of these proteins in MSCs from AML recipient mice using qPCR. The results showed that Angptl7 mRNA levels were upregulated in AML BMSCs (Figure 4C). We performed enzyme-linked immunosorbent assay (ELISA), western blot, and qPCR assays to assess the level of ANGPTL7 in BM fluid and MSCs derived from patients with AML and found that ANGPTL7 levels were significantly increased in AML samples compared with those in healthy individuals (Figure 4D-E; supplemental Figure 7D). Patients with AML subtype M2 or M5 with high ANGPTL7 expression have a significantly worse overall survival than patients with low ANGPTL7 expression39 (supplemental Figure 7E).
Id1 enhances Angptl7 release from BMSCs to promote AML progression. (A) The proteomics analysis strategy of BMSF from Id1+/+ and Id1−/− AML recipients. (B) The heat map analysis of differentially expression proteins in tandem mass tag–based proteomic analysis of BMSF from wild-type and Id1−/− AML mice. (C) The qPCR analysis of the differentially expressed proteins in BMSF proteomics. (D) The ELISA analysis of ANGPTL7 levels in the BM fluid derived from healthy individuals (n = 3) and patients with AML (M2, n = 2; M5, n = 2). (E) The western blotting analysis of ANGPTL7 levels in the MSCs derived from healthy individuals (n = 2) and patients with AML (M2, n = 2; M5, n = 2). (F) The ELISAs showed that ID1 knockout in HS-5 cells significantly decreases the ANGPTL7 protein level in CM. Overexpression of HA-ID1 in HS-5 cells significantly upregulates the level of ANGPTL7 in CM. (G) The MTT assays showed that ANGPTL7 recombinant protein significantly promotes the proliferation of Kasumi-1 and THP-1 cells. (H) CFU assays analyzing the AE9a LSKs or MLL-AF9 L-GMP cells. Average number of colonies generated from 800 cells (n = 3). (I-K) Cell cycle analysis on Kasumi-1 and THP-1 cells with vehicle or ANGPTL7 during coculture with ID1+/+ or ID1−/− HS-5 cells (n = 3). (L-N) Cell cycle analysis on AE9a-LSKs and MLL-AF9-L-GMP cells with vehicle or Angptl7 during coculture with Id1ctrl or Id1kd MS-5 cells (n = 3). (O) The western blot analysis of lentivirus-mediated ID1 silencing in patient-derived AML subtype M2 and M5 MSCs. Anti-ID1, anti-SP1, anti-ANGPTL7, and anti-GAPDH antibodies were used. (P-Q) The MTT proliferation analysis on patient-derived AML subtype M2 and M5 CD34+ leukemia blast cells with vehicle or ANGPTL7 treatment during coculture with matched ID1+/+ or ID1−/− MSCs. ∗P < .05; ∗∗P < .01; ∗∗∗P < .005; ∗∗∗∗P < .001. FC, fold change; WT, wild-type.
Id1 enhances Angptl7 release from BMSCs to promote AML progression. (A) The proteomics analysis strategy of BMSF from Id1+/+ and Id1−/− AML recipients. (B) The heat map analysis of differentially expression proteins in tandem mass tag–based proteomic analysis of BMSF from wild-type and Id1−/− AML mice. (C) The qPCR analysis of the differentially expressed proteins in BMSF proteomics. (D) The ELISA analysis of ANGPTL7 levels in the BM fluid derived from healthy individuals (n = 3) and patients with AML (M2, n = 2; M5, n = 2). (E) The western blotting analysis of ANGPTL7 levels in the MSCs derived from healthy individuals (n = 2) and patients with AML (M2, n = 2; M5, n = 2). (F) The ELISAs showed that ID1 knockout in HS-5 cells significantly decreases the ANGPTL7 protein level in CM. Overexpression of HA-ID1 in HS-5 cells significantly upregulates the level of ANGPTL7 in CM. (G) The MTT assays showed that ANGPTL7 recombinant protein significantly promotes the proliferation of Kasumi-1 and THP-1 cells. (H) CFU assays analyzing the AE9a LSKs or MLL-AF9 L-GMP cells. Average number of colonies generated from 800 cells (n = 3). (I-K) Cell cycle analysis on Kasumi-1 and THP-1 cells with vehicle or ANGPTL7 during coculture with ID1+/+ or ID1−/− HS-5 cells (n = 3). (L-N) Cell cycle analysis on AE9a-LSKs and MLL-AF9-L-GMP cells with vehicle or Angptl7 during coculture with Id1ctrl or Id1kd MS-5 cells (n = 3). (O) The western blot analysis of lentivirus-mediated ID1 silencing in patient-derived AML subtype M2 and M5 MSCs. Anti-ID1, anti-SP1, anti-ANGPTL7, and anti-GAPDH antibodies were used. (P-Q) The MTT proliferation analysis on patient-derived AML subtype M2 and M5 CD34+ leukemia blast cells with vehicle or ANGPTL7 treatment during coculture with matched ID1+/+ or ID1−/− MSCs. ∗P < .05; ∗∗P < .01; ∗∗∗P < .005; ∗∗∗∗P < .001. FC, fold change; WT, wild-type.
We knocked out or overexpressed ID1 in HS-5 cells, and ELISA analysis of the conditioned medium showed that ANGPTL7 was significantly downregulated in ID1−/− HS-5 cells and highly expressed in HS-5 cells overexpressing ID1 (Figure 4F). To demonstrate that ANGPTL7 promotes AML cell proliferation, Kasumi-1 and THP-1 cells were treated with or without the human recombinant ANGPTL7 protein. The MTT analysis shows that Kasumi-1 and THP-1 cells grow faster in the medium containing ANGPTL7 than cells in the control group (Figure 4G). Moreover, we constructed a plasmid containing the entire encoding sequence of murine Angptl7 with a C-terminal 3× Flag label in a eukaryotic expression vector. MS-5 supernatants contained Angptl7-Flag fusion protein and was collected using FLAG affinity resin and used for in vitro and in vivo experiments (supplemental Figure 7F). The colony formation assays showed that the size and number of colonies generated by AE9a-expressing LSKs or MLL-AF9–expressing L-GMP cells, which were treated with Angptl7 protein, were significantly increased (Figure 4H; supplemental Figure 7G). To further examine the possibility that ANGPTL7 is regulated by ID1 in the BMM, we performed in vitro coculture assays. Flow analysis showed that supplementation of ANGPTL7 significantly restored G0/G1 cell cycle arrest in AML cells cocultured with ID1−/− HS-5 cells (Figure 4I-K). Similarly, flow analysis showed a significant reduction of G0/G1 cell cycle arrest in AE9a-expressing LSKs or MLL-AF9 L-GMP cells when Angptl7 was added to MS-5 or OP9 cells with the knockdown of Id1 (Figure 4L-N; supplemental Figure 7H-I).
To better investigate the function of ID1 in AML-MSCs, we knocked out ID1 in MSCs derived from patients with AML subtype M2 or M5 and found that deletion of ID1 significantly reduced ANGPTL7 levels (Figure 4O; supplemental Figure 7J). We performed MTT and colony formation assays on CD34+ leukemia blast cells cocultured with matched ID1+/+ or ID1−/− MSCs derived from patients with AML (supplemental Figure 7K). The results showed that ID1−/− AML-derived MSCs significantly inhibits the proliferation and colony-forming capacity of CD34+ leukemia blast cells, which was significantly restored via ANGPTL7 treatment (Figure 4P-Q; supplemental Figure 7L-M). Together, these results suggest that ID1 regulates ANGPTL7 in MSCs and promotes the development of leukemia.
ID1-mediated SP1 protein level is essential for the transcription of ANGPTL7
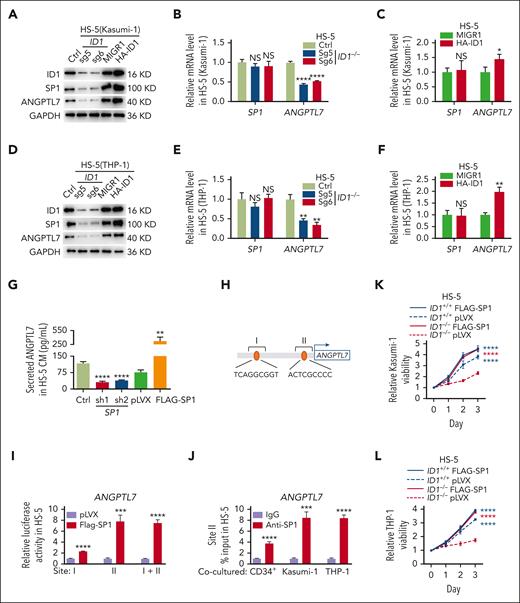
SP1, a ubiquitously expressed transcription factor that binds GC-rich cis elements, reportedly upregulates ANGPTL7 in human glaucoma.59 Interestingly, we found that in HS-5 cells cocultured with Kasumi-1 cells, the deletion of ID1 reduced the protein level but not the mRNA level of SP1. Conversely, ID1 overexpression increased the SP1 protein level but not the mRNA level (Figure 5A-C). Knocking down of ID1 also decreased the protein level but not the mRNA level of SP1 in HS-5 cells cocultured with THP-1 cells and overexpression of ID1 increased the SP1 protein level but not the mRNA level (Figure 5D-F). However, knockdown or overexpression of SP1 in HS-5 cells had little effect on ID1 protein (supplemental Figure 8A). Thus, we hypothesize that ID1 regulates the expression of SP1 at the posttranscriptional level to control the transcription of ANGPTL7.
ID1-mediated SP1 protein levels are essential for Angptl7 transcription. (A-C) HS-5 cells were transduced with indicated sgRNAs and overexpression plasmids. Western blott and qPCR analyses were performed on these HS-5 cells after 4 days of coculture with Kasumi-1 cells. (D-F) HS-5 cells were transduced with indicated sgRNAs and overexpression plasmids. Western blot and qPCR analyses were performed on these HS-5 cells after 4 days of coculture with THP-1 cells. (G) The ELISAs showed that SP1 knockdown restricts ANGPTL7 expression and that ANGPTL7 was significantly upregulated by SP1 overexpression in HS-5 cells CM. (H) PROMO predicts the SP1 binding site on the ANGPTL7 promoter. (I) The dual-luciferase reporter assay showed that SP1 binds to site II to upregulate luciferase activity. (J) Chromatin immunoprecipitation assays showed that ANGPTL7 promoter DNA fragments were immunoprecipitated by SP1 antibodies in contrast to immunoglobulin G (IgG) in HS-5 cells cocultured with CD34+ HSPCs, Kasumi-1, or THP-1 cells. (K-L) The MTT assays showed that overexpression of SP1 in ID1−/− HS-5 cells significantly promote the proliferation of Kasumi-1 and THP-1 cells. ∗P < .05; ∗∗P < .01; ∗∗∗P < .005; ∗∗∗∗P < .001.
ID1-mediated SP1 protein levels are essential for Angptl7 transcription. (A-C) HS-5 cells were transduced with indicated sgRNAs and overexpression plasmids. Western blott and qPCR analyses were performed on these HS-5 cells after 4 days of coculture with Kasumi-1 cells. (D-F) HS-5 cells were transduced with indicated sgRNAs and overexpression plasmids. Western blot and qPCR analyses were performed on these HS-5 cells after 4 days of coculture with THP-1 cells. (G) The ELISAs showed that SP1 knockdown restricts ANGPTL7 expression and that ANGPTL7 was significantly upregulated by SP1 overexpression in HS-5 cells CM. (H) PROMO predicts the SP1 binding site on the ANGPTL7 promoter. (I) The dual-luciferase reporter assay showed that SP1 binds to site II to upregulate luciferase activity. (J) Chromatin immunoprecipitation assays showed that ANGPTL7 promoter DNA fragments were immunoprecipitated by SP1 antibodies in contrast to immunoglobulin G (IgG) in HS-5 cells cocultured with CD34+ HSPCs, Kasumi-1, or THP-1 cells. (K-L) The MTT assays showed that overexpression of SP1 in ID1−/− HS-5 cells significantly promote the proliferation of Kasumi-1 and THP-1 cells. ∗P < .05; ∗∗P < .01; ∗∗∗P < .005; ∗∗∗∗P < .001.
We performed ELISA assays on conditioned medium of HS-5 cells 48 hours after either knocking out or overexpression of SP1. The results showed that SP1 knockdown restrained ANGPTL7 level, and SP1 overexpression increased ANGPTL7 level (Figure 5G). Given the crucial role of SP1 in the expression of ANGPTL7, we performed luciferase reporter and chromatin immunoprecipitation qPCR assays to test for SP1 binding to the ANGPTL7 promoter. Transcription factors that potentially bind to the promoter region of ANGPTL7 were predicted using PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3), and 2 potential binding sites were predicted (Figure 5H). A dual-luciferase reporter assay was applied to detect the relationship between SP1, and both sites showed that luciferase activity was significantly upregulated because of the adoption of Flag-SP1, compared with the empty vector in promoter site II of ANGPTL7. Hence, SP1 was supposed to target this region of ANGPTL7 promoter in HS-5 cells (Figure 5I). Furthermore, the results of chromatin immunoprecipitation assays indicated that DNA fragments at the ANGPTL7 promoter were obviously immunoprecipitated by the anti-SP1 antibody in comparison with the immunoglobulin G control in HS-5 cells after coculturing with CD34+ HSPCs, Kasumi-1, or THP-1 cells for 4 days (Figure 5J). The result of MTT assays showed that overexpression of SP1 in ID1−/− HS-5 cells significantly rescued the proliferation of Kasumi-1 and THP-1 cells (Figure 5K-L). ID1 knockout significantly reduced SP1 protein levels in HS-5 cells cocultured with AML cells, followed by a suppression of ANGPTL7 transcription.
The interaction between ID1 and RNF4 suppresses SP1 ubiquitination
Because ID1 is important for the maintenance of the SP1 protein, we simultaneously caused the overexpression of FLAG-SP1 in HA-ID1–expressing HS-5 cells and examined their ability to interact with stably expressed exogenous HA-ID1 using coimmunoprecipitation and western blot assays. The results showed that there is no direct interaction between SP1 and ID1 (supplemental Figure 8B-C). To investigate how ID1, a negative–DNA-binding factor, modulates SP1 protein levels, we performed an ID1-interactome analysis using mass spectrometry. Whole-cell lysates of MS-5 cells cocultured for 4 days with MLL-AF9–expressing leukemia cells were immunoprecipitated using anti-HA antibodies, and an analysis of the silver nitrate staining showed that HA-Id1 was immunoprecipitated. The Rnf4, ubiquitin E3 ligase of Sp1,60,61 interacting with HA-Id1 was identified (Figure 6A; supplemental Table 3). The pCMV-RNF4-FLAG plasmid were simultaneously transfected into HA-ID1–expressing HS-5 cells and the control cells, and coimmunoprecipitation assays confirmed the interaction between HA-ID1 and RNF4 (Figure 6B-C). We performed GST pull-down assays, which showed a direct interaction between ID1 and RNF4 (Figure 6D). Next, expression of RNF4 and SP1 were examined using qPCR and western blot analysis, respectively, in HS-5 cells transduced with shRNA against RNF4 or a control shRNA. The downregulation of RNF4 significantly upregulated SP1 protein but did not affect ID1 protein (Figure 6E). To determine whether ID1–RNF4 interaction affects SP1 protein, we performed GST pull-down assay and examined a significant reduction in the amount of GST-RNF4 binding SP1 in the presence of HA-ID1 (Figure 6F). We performed ubiquitination assays and examined whether ID1 inhibited SP1 ubiquitination via RNF4. SP1, HA-ID1, RNF4-FLAG, and MYC-ubiquitin were cotransfected into 293T cells. We then performed coimmunoprecipitation assays in these cells using anti-SP1 antibodies and observed a significant reduction in the ubiquitination of the SP1 protein. This result suggests that RNF4 is not an E3 ubiquitin ligase of ID1, and that their interaction inhibits SP1 ubiquitination (Figure 6G; supplemental Figure 8D). To examine whether ID1–RNF4 interaction could influence AML progression, we sought to knock down both ID1 and RNF4 in HS-5 cells and performed coculture assays of Kasumi-1 or THP-1 cells with these HS-5 cells for 3 days (supplemental Figure 8E). The results of MTT assays showed that knock down RNF4 can rescue ID1 deficiency–disrupted nonautonomous cell support for Kasumi-1 and THP-1 cells (supplemental Figure 8F-G).
The interaction between ID1 and RNF4 suppresses SP1 ubiquitination. (A) The silver nitrate staining results of the immunoprecipitation HA-Id1 interactome. (B-C) Both HA and FLAG coimmunoassays showed significant interactions between HA-ID1 and RNF4-FLAG. (D) ID1 and RNF4 interact with each other in vitro. Both purified GST-RNF4 and GST-ID1 pull-down assays showed significant interactions with His-ID1 and His-RNF4. (E) The levels of SP1 protein, but not ID1, were significantly reduced in HS-5 cells transduced with shRNA against RNF4. (F) ID1 inhibits the RNF4-SP1 interaction in vitro. Purified GST-RNF4, HA-ID1, and FLAG-SP1 were used for GST pull-down assays, and anti-GST, anti-HA, and anti-FLAG antibodies were used for western blotting. (G) SP1, HA-ID1, RNF4-FLAG, and MYC-ubiquitin were cotransfected into 293T cells. Anti-SP1 immunoprecipitation assays have shown that HA-ID1 significantly inhibits SP1 ubiquitination via RNF4. (H) Schematic representation of the ID1 and RNF4 truncations. (I) The anti-FLAG immunoprecipitation assay showed a significant RNF4-FLAG interaction with the ID1 C-terminal. (J) Anti-HA immunoprecipitation assay showed significant interaction of HA-ID1 with the RNF4 SIM domain. (K) HS-5 cells with stable expression of truncated ID1 were transiently transfected with either RNF4 or empty vectors. Western blot analysis of SP1, HA, and FLAG-related proteins. (L-M) MTT assays showed significant inhibition of the proliferation in Kasumi-1 and THP-1 cells cocultured with HS-5 cells expressing RNF-FLAG and HA-ID1 C del. ∗P < .05; ∗∗P < .01; ∗∗∗P < .005; ∗∗∗∗P < .001.
The interaction between ID1 and RNF4 suppresses SP1 ubiquitination. (A) The silver nitrate staining results of the immunoprecipitation HA-Id1 interactome. (B-C) Both HA and FLAG coimmunoassays showed significant interactions between HA-ID1 and RNF4-FLAG. (D) ID1 and RNF4 interact with each other in vitro. Both purified GST-RNF4 and GST-ID1 pull-down assays showed significant interactions with His-ID1 and His-RNF4. (E) The levels of SP1 protein, but not ID1, were significantly reduced in HS-5 cells transduced with shRNA against RNF4. (F) ID1 inhibits the RNF4-SP1 interaction in vitro. Purified GST-RNF4, HA-ID1, and FLAG-SP1 were used for GST pull-down assays, and anti-GST, anti-HA, and anti-FLAG antibodies were used for western blotting. (G) SP1, HA-ID1, RNF4-FLAG, and MYC-ubiquitin were cotransfected into 293T cells. Anti-SP1 immunoprecipitation assays have shown that HA-ID1 significantly inhibits SP1 ubiquitination via RNF4. (H) Schematic representation of the ID1 and RNF4 truncations. (I) The anti-FLAG immunoprecipitation assay showed a significant RNF4-FLAG interaction with the ID1 C-terminal. (J) Anti-HA immunoprecipitation assay showed significant interaction of HA-ID1 with the RNF4 SIM domain. (K) HS-5 cells with stable expression of truncated ID1 were transiently transfected with either RNF4 or empty vectors. Western blot analysis of SP1, HA, and FLAG-related proteins. (L-M) MTT assays showed significant inhibition of the proliferation in Kasumi-1 and THP-1 cells cocultured with HS-5 cells expressing RNF-FLAG and HA-ID1 C del. ∗P < .05; ∗∗P < .01; ∗∗∗P < .005; ∗∗∗∗P < .001.
We transfected the full length or truncated plasmid of HA-ID1 and FLAG-RNF4 into 293T cells and observed an interaction between HA-ID1 C-terminal and FLAG-RNF4-SIM in these cells by using coimmunoprecipitation assays (Figure 6H-J). The SIM domain is reportedly essential for RNF4 recruitment and promotion of SP1 ubiquitination.62,63 Thus, the interaction between ID1 and RNF4 acts as a SIM domain blocker and weakens the ubiquitination of SP1. We constructed HS-5 cells that stably express ID1 truncates and transiently transfected FLAG-RNF4 or the empty vector (Figure 6K). We performed coculture assays of Kasumi-1 or THP-1 cells with these HS-5 cells for 4 days. The results of MTT assays showed that the C terminus of ID1 is required for its interaction with RNF4 (Figure 6L-M). ID1 competes with SP1 for binding to the SIM domain of RNF4, which inhibits SP1 ubiquitination and contributes to AML progression nonautonomously.
Intra-BM transfusion of Angptl7 significantly accelerated AML progression in recipients deficient in Id1
To confirm that ID1 modulates ANGPTL7 in the AML BMM to maintain LSCs and affect AML progression in vivo, we attempted an intra-BM injection based on the findings of other investigators.64 We initially attempted systemic administration of Angptl7 via the tail vein and did not observe that ANGPTL7 was as potent in vivo as at the cellular level, presumably because ANGPTL7 is unstable in peripheral circulation. For these reasons, intrafemoral injections were attempted. After Angptl7 treatment, the survival of Angptl7-treated Id1−/− leukemia-bearing mice was significantly shortened in AE9a- and MLL-AF9–derived leukemia (Figure 7A-B). Four weeks after transplantation, the white blood cell counts of the Id1−/− recipients without the treatment of Angptl7 were significantly lower than those of other groups (supplemental Figure 9A-B). Splenomegaly was prominently observed in the Id1+/+ mice with or without the treatment of Angptl7 and less in the Id1−/− recipients without the treatment of Angptl7 (supplemental Figure 9C-E). Flow cytometry analysis showed that the BM, spleen, and peripheral blood cells of Id1−/− recipients without the treatment of Angptl7 contained a lower percentage of GFP+c-Kit+ leukemia blast cells and a significantly decreased percentage of L-GMP cells in MLL-AF9–driven leukemia than the cells of Id1+/+ mice (Figure 7C-H). Moreover, the percentage of AE9a-expressing GFP+Mac-1− and MLL-AF9–expressing GFP+Mac-1+ cells in the BM, spleen, and peripheral blood cells of Id1−/− recipients without Angptl7 treatment revealed significant reduction (supplemental Figure 9F-I). In summary, our data suggest that Id1 regulates Angptl7 to promote the development of leukemia.
Intra-BM transfusion of Angptl7 significantly accelerated AML progression in Id1-deficient recipient mice. (A-B) Id1−/− recipient mice without Angptl7 injection showed significantly longer survival time than other groups after FLT or BMT. (C-F) The frequency of GFP+c-Kit+ leukemia blast cells was decreased in the BM, spleen, and peripheral blood (PB) of Id1−/− recipients without Angptl7 injection after FLT and BMT (n = 8). (G-H) Id1−/− recipients without Angptl7 injection showed a decrease in the frequency of L-GMP cells in the BM after BMT (n = 8). ∗P < .05; ∗∗P < .01; ∗∗∗P < .005; and ∗∗∗∗P < .001.
Intra-BM transfusion of Angptl7 significantly accelerated AML progression in Id1-deficient recipient mice. (A-B) Id1−/− recipient mice without Angptl7 injection showed significantly longer survival time than other groups after FLT or BMT. (C-F) The frequency of GFP+c-Kit+ leukemia blast cells was decreased in the BM, spleen, and peripheral blood (PB) of Id1−/− recipients without Angptl7 injection after FLT and BMT (n = 8). (G-H) Id1−/− recipients without Angptl7 injection showed a decrease in the frequency of L-GMP cells in the BM after BMT (n = 8). ∗P < .05; ∗∗P < .01; ∗∗∗P < .005; and ∗∗∗∗P < .001.
Discussion
Although clinical governance of AML has improved significantly over the past few years, significant challenges, such as the high molecular diversity and complexity found in AML, the variable evolution of the disease, and the course of treatment, all of which partly depend on the BMM, remain.65,66
ID1 is a key transcriptional regulator in LSCs previously identified by us; however, the function of ID1 in the AML BMM has not been identified. In this study, we identified an unexpected effect of ID1 on SP1 protein levels, suggesting that reduced SP1 degradation may be a key mechanism of ID1, given that the overexpression of SP1 in ID1-deficient mesenchymal cells can reverse the proliferation of cocultured AML cells. Because ID1 does not interact directly with SP1, we performed an ID1 interaction analysis using mass spectrometry, and RNF4, the ubiquitin E3 ligase of SP1,60,61 was identified. We have determined that ID1 interacts with RNF4 through its C-terminal region and blocks the RNF4 SIM domain, consistent with the reduction in SP1 ubiquitination induced by RNF4 knockdown. Indeed, the overexpression of ID1 C del and RNF4 in mesenchymal cells significantly modulates SP1 protein levels and suppresses the proliferation of cocultured AML cells compared with other ID1 truncations. Knocking down RNF4 can rescue SP1 degradation induced by ID1 deficiency in HS-5 cells.
SP1 plays an important role in glaucoma by upregulating ANGPTL7.59,67,68 In Id1-knockout recipient AML mice, levels of Angptl7 protein in the BMSF were significantly downregulated, and survival was significantly longer, suggesting that Id1-mediated Sp1 protein levels may be essential for Angptl7 transcription in AML progression. Our study showed that overexpression of SP1 in ID1-deleted mesenchymal cells significantly rescued cocultured AML cell proliferation. Targeting ID1 may represent a potential treatment for AML by reducing levels of the SP1 protein and its target gene, Angptl7.
Altogether, we report here that ID1 interacts directly with RNF4, attenuates SP1 ubiquitination, and contributes to the expression of ANGPTL7 in AML-MSCs. More importantly, as a key transcriptional regulator in MSCs that supports the noncellular autonomy of leukemia cells identified here, ID1 controls the initiation and progression of leukemia in a cell-autonomous role, suggesting the importance of ID1 as a therapeutic target for AML.
Acknowledgments
The authors thank Y. Zhai, X. Miao, L. Qiu, S. Yan, P. Zhou, K. Wang, Z. Wong, Y. Bu, K. Pan, J. Wu, and X. Yan from the Institutional Center for Shared Technologies and Facilities of SINH, CAS, for technical assistance. The authors also thank Dangsheng Li for critical reading and helpful comments on the manuscript.
This work was supported by the National Key Research and Development Plan of China (2018YFA0800203 and 2018YFA0107202 [L.W.]); the National Natural Science Foundation of China General Program (81970150 and 82170156 [L.W.]); Shanghai Science and Technology Innovation Action Plan Excellent Academic/Technical Leader Program (Youth) (21XD1424500 [L.W.]).
Authorship
Contribution: L.W. conceived the study; L.W. and M.-Y.F. designed the study, analyzed the data, and wrote the paper; Q.-L.Z, L.-Y.W., J.Y., S.S., C.-K.C., and X.-J.S. analyzed the data and contributed new tools; K.X., F.D., D. Huang., M.-H.H., C.Z., J.-C.J., and R.-J.W. acquired patient samples; H.L. contributed to acquiring the cell line; M.-Y.F., Y.W., B.-H.C., X.-Y.L., Z.-J.L., C.-L.H., P.L., J.-C.W., P.-C.Y., S.-B.C., C.-H.X., B.-Y.C., Y.-L.J., N.L., C.Z., D. Hou., X.-C.C., Y.-Y.R., C.-H.D., J.-Y.Z., and L.-j.Z. performed the study; B.-H.C. and C.-H.X. performed the bioinformatics analysis; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lan Wang, Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China; e-mail: lwang@sinh.ac.cn.
References
Author notes
The data sets generated in this study have been deposited in Gene Expression Omnibus database (accession number GSE219049).
Data are available on request from the corresponding author, Lan Wang (lwang@sinh.ac.cn).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.