In this issue of Blood, Heger and colleagues describe a novel tool for predicting clinical outcome in central nervous system (CNS) lymphomas that is based on clinical features, radiographic response to induction, and circulating tumor DNA (ctDNA) detection.1
Lymphomatous involvement of the CNS is a much-feared clinical scenario with an uncertain prognosis.2 Whether it represents a primary CNS lymphoma (PCNSL) or occurs in the context of a systemic aggressive lymphoma at initial diagnosis or relapse, outcomes are relatively poor. Even today, clinical features such as age and performance status remain the basis for risk stratification.3,4 High-dose methotrexate-containing regimens that culminate in myeloablative chemotherapy and autologous stem cell transplant are associated with the best outcomes in young fit patients and may be curative. Older patients or those with comorbidities are often ineligible for intensive strategies and are consolidated with radiotherapy, with increased risk of cognitive impairment. Response assessments rely on magnetic resonance imaging (MRI), which may not discriminate between residual scar and persistent disease. Consequently, new tools that can reliably distinguish risk subgroups and define tumor response are needed to inform treatment intensity and avoid unnecessary toxicity in these often frail patients.
In systemic diffuse large B-cell lymphoma, ctDNA is a promising indicator of residual disease that is under intensive investigation.5 In CNS lymphomas, the very low levels of ctDNA present in plasma and difficulties in obtaining primary tissue and serial cerebrospinal fluid (CSF) specimens for study have challenged investigators working to apply a similar strategy to patients with lymphoma limited to the CNS.6 Both single-gene polymerase chain reaction assays for the detection of MYD88L265P, the most frequent mutation in PCNSL, and next-generation sequencing (NGS)-based approaches have been applied to CSF and plasma with variable results. In a recent study by Mutter and colleagues, an ultrasensitive high-throughput sequencing approach detected ctDNA in 61 of 78 plasma specimens and 24 of 24 CSF samples collected from patients with PCNSL or isolated secondary CNSL.7 Notably, detection of plasma ctDNA at diagnosis, during treatment, or at completion of therapy, representing residual disease, was associated with poor outcomes.
Heger et al here report on development of a robust prognostic model based on an ultrasensitive ctDNA sequencing approach in samples from 67 patients with CNSL (58 with PCNSL; 9 with secondary CNSL) that is tumor agnostic. This method allowed for the detection of CNS lymphoma-derived mutations in plasma ctDNA with high concordance to CSF and tumor tissue. Consistent with the findings of Mutter et al, patients with PCNSL without detectable ctDNA in plasma at baseline had a nonsignificant trend to favorable outcomes. More importantly, the persistence of “peripheral residual disease” (a term used by the authors and defined by detectable levels of ctDNA in plasma samples collected during or after treatment) was associated with a near significant trend toward impaired failure-free survival. Peripheral residual disease seems to be a reliable parameter of incomplete tumor regression and may be a suitable tool to drive intensification/consolidation strategies in these patients.
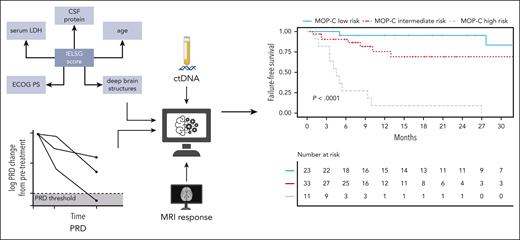
Heger and colleagues should be commended for their efforts to improve the accuracy and to expand the value of the International Extranodal Lymphoma Study Group (IELSG) score as a prognostic index for PCNSL patients. Incorporating a combination of baseline parameters (ie, IELSG clinical features and ctDNA levels), MRI results, and sequential measurements of ctDNA, they built a prognostic model applicable to patients treated with high-dose methotrexate-containing induction. In contrast to most prognostic indices that predict outcome from the outset, the model proposed by Heger et al, called Molecular Prognostic Index for CNS lymphomas (MOP-C), provides an “interim” assessment of prognosis that has the potential to more accurately identify individuals destined to fail conventional therapy, with 2-year failure-free survival rates of 95%, 69%, and 9%, respectively, for patients at low, intermediate, and high risk (see figure).
CtDNA sequencing was applied to plasma and CSF samples using a targeted gene panel. Peripheral residual disease (PRD) was developed as a novel biomarker in CNSL and shown to be predictive of outcomes. Integration of dynamically assessed clinical and molecular features resulted in development and validation of MOP-C—a tool with high predictive value in CNSL. ECOG PS, Eastern Cooperative Oncology Group performance status; IELSG, International Extranodal Lymphoma Study Group. Created with BioRender.com. See Figure 1 in the article by Heger et al that begins on page 522.
CtDNA sequencing was applied to plasma and CSF samples using a targeted gene panel. Peripheral residual disease (PRD) was developed as a novel biomarker in CNSL and shown to be predictive of outcomes. Integration of dynamically assessed clinical and molecular features resulted in development and validation of MOP-C—a tool with high predictive value in CNSL. ECOG PS, Eastern Cooperative Oncology Group performance status; IELSG, International Extranodal Lymphoma Study Group. Created with BioRender.com. See Figure 1 in the article by Heger et al that begins on page 522.
The encouraging results reported by Heger et al deserve to be addressed in larger numbers of patients in prospective clinical trials. A phase 2 trial incorporating MOP-C (NCT05583071) is now underway. This is an important issue, especially if we consider that some comparisons did not reach significance due to the small size of analyzed subgroups. Some questions remain open for future studies. According to prior PCNSL studies, ctDNA analyses performed on CSF samples produced more reliable results than analyses on plasma. Likewise, in the study by Heger et al, the share of total cell-free DNA originating from the tumor was increased in CSF as compared with plasma samples, but unfortunately, only 9 CSF samples were studied. Thus, the most suitable sample for ctDNA analysis remains to be defined. The significance of noncoding mutations in plasma samples should also be clarified, especially in the 12% of cases where this was the only recorded abnormality. That some “IELSG variables” (ie, performance status [PS], lactate dehydrogenase [LDH] serum level, CSF protein concentration) change remarkably during treatment should be considered. A dynamic assessment of MOP-C scores at several time points (eg, baseline, midtreatment, and posttreatment) may be a better predictor of outcomes than a single assessment.
Although ctDNA promises to vastly improve risk and response assessments in CNS lymphomas, paving the way for a wide range of novel study designs, there are significant challenges to be overcome. The methods, gene panels, and algorithms reported in lymphoma studies are diverse, with varied sensitivities and allelic mutation rates. Thus, independent validation and standardization of available methods remain important goals. The optimal timing of ctDNA assessments in PCNSL also remains to be determined. Reliable and affordable approaches to detecting ctDNA will be required for widespread integration into treatment algorithms.
Conflict-of-interest disclosure: J.N.W. has received research funding and honoraria from Merck. A.J.M.F. declares no competing financial interest.