Abstract
Protein S deficiency is a known risk factor for thrombosis. The coexistence of phenotypic type I (reduction in total and free antigen) and type III (reduction in free antigen only) protein S deficiencies in 14 of 18 families was recently reported. We investigated the cause of this phenotypic variation in the largest of these families (122 family members, including 44 affected individuals) using both molecular genetic and phenotypic analysis. We have identified a sole causative mutation (Gly295Val) in three family members representative of the variable phenotype. Complete cosegregation of the mutation with reduced free protein S antigen levels was found, regardless of the total antigen level. Analysis of phenotypic data showed high correlations between total protein S antigen and age in both normal and protein S–deficient family members, irrespective of gender. Free protein S antigen levels were not influenced by age, a finding explained by an association between β-chain containing C4b-binding protein (C4bBP-β+) antigen levels and age. We propose that the identified Gly295Val mutation causes quantitative, or type I, protein S deficiency, and that as age increases the total protein S antigen level normalizes with respect to the reference plasma pool, giving rise to a type III protein S–deficient phenotype.
PROTEIN S ACTS AS A nonenzymic cofactor to activated protein C (APC) in the degradation of activated factors V and VIII in the protein C anticoagulant system (reviewed in Dahlbäck1 ). It has also been shown to inhibit both the prothrombinase complex2-4 and the factor X activating complex5 independently of APC. The importance of the anticoagulant role that protein S plays is evident in individuals with homozygous protein S deficiency, who suffer from severe purpura fulminans from birth.6,7 Heterozygous protein S deficiency is also thought to be a risk factor for venous thrombosis8 and is found in 1% to 7% of patients with unexplained thrombosis.9-12
The plasma concentration of protein S is approximately 20 to 25 mg/L,13 of which a proportion is noncovalently bound to C4b-binding protein (C4bBP),14 a component of the classic complement pathway. C4bBP consists of seven or eight α-chains that bind to C4b and either one or no β-chain that contains the binding site for protein S.15,16 The amount of free protein S antigen has been shown to be equal to the molar excess of total protein S over β-chain containing C4bBP (C4bBP-β+),17 approximately 30% to 40% of total protein S in normal individuals. Only free protein S is capable of acting as a cofactor to APC.18 The protein S domain structure is largely homologous to other vitamin K–dependent proteins at the amino terminus, whereas the carboxy-terminal portion has been found to be homologous to sex hormone–binding globulin (SHBG).19 Two genes for human protein S have been identified20-22 after the cDNA was cloned.23-25 One is the active gene (PROS1), and the other contains multiple frameshifts and stop codons and has been classified as a pseudogene.
Diagnosis of protein S deficiency is assessed using a variety of plasma-based assays for both activity and antigen; both total and free protein S antigen levels can be measured. Using these measurements, protein S deficiency can be classified into three phenotypic subtypes (using the classification proposed at the meeting of the Scientific Subcommittee of the International Society on Thrombosis and Haemostasis, Munich, Germany, 1991). Type I protein S deficiency is associated with a reduction in activity and total and free antigen levels (quantitative deficiency), type II with normal total and free antigen levels and reduced activity (functional variant), and type III with normal total protein S antigen levels and reduced free antigen levels and activity. One of the problems encountered in the diagnosis of protein S deficiency is the presence of a large overlap in antigen levels between normal and heterozygous individuals.26
Previously, it has been reported that type I and type III protein S deficiencies coexisted within 14 of 18 protein S–deficient families,27 which led to the proposal that these subtypes are phenotypic variants of the same genetic disease. The entire coding region and intron/exon boundaries of PROS1 in selected members of a large family that displayed phenotypic variation have now been analyzed. This has enabled us to reassess the plasma phenotype of protein S deficiency, based on genetically determined diagnosis, and to propose an explanation for the variable protein S phenotype.
MATERIALS AND METHODS
Patients and plasma phenotype determination.The protein S–deficient kindred involved in this study has been reported previously as family no. 1.27 Of 122 family members available for both phenotypic and genotypic analysis, 30 had venous thrombosis. All had been investigated previously for the factor V Arg506Gln mutation,28 which is associated with APC resistance.29 Using the previously described method,30 31 a single family member (VI-5) was heterozygous at this site. The family tree is illustrated in Fig 1.
Family relationship of germline individuals in the protein S–deficient kindred currently under investigation. Additional untested relatives are shown if they were siblings or ancestors of tested family members. All protein S–deficient individuals tested had the Gly295Val mutation. Protein S deficiency subclassification for nonanticoagulated affected family members is as used for genetic analysis (type I, III, and I/III phenotypes). The 3 individuals selected for genetic analysis are indicated with an asterisk (IV-30, V-34, and VI-18). Investigated family members who had venous thrombosis are indicated with a diamond. The single family member who was heterozygous for the factor V Arg506Gln mutation is indicated with a cross.
Family relationship of germline individuals in the protein S–deficient kindred currently under investigation. Additional untested relatives are shown if they were siblings or ancestors of tested family members. All protein S–deficient individuals tested had the Gly295Val mutation. Protein S deficiency subclassification for nonanticoagulated affected family members is as used for genetic analysis (type I, III, and I/III phenotypes). The 3 individuals selected for genetic analysis are indicated with an asterisk (IV-30, V-34, and VI-18). Investigated family members who had venous thrombosis are indicated with a diamond. The single family member who was heterozygous for the factor V Arg506Gln mutation is indicated with a cross.
Methods for determination of plasma protein S and C4bBP-β+ antigen have been described previously.27 In brief, total and free protein S antigen levels were measured by radioimmunoassay,32 and C4bBP-β+ antigen level was measured using an enzyme-linked immunosorbent assay.33 Reference ranges for free and total protein S antigen levels were 56 to 182 nmol/L and 219 to 407 nmol/L, respectively. Individuals who were receiving oral anticoagulant treatment at the time of testing were diagnosed by comparison to an anticoagulated control group (treatment unrelated to protein S deficiency). The reference range for free protein S antigen in this group was 16 to 91 nmol/L. Individuals were classified as protein S–deficient if the free protein S antigen level was less than the lower limit of the reference range (n = 44).
Protein S–deficient individuals were provisionally subclassified for the purpose of genetic analysis. The limits used were as follows: type I protein S–deficient, total protein S antigen levels 188 nmol/L or less (n = 12); type III, total protein S antigen 250 nmol/L or greater (n = 4); and type I/III, total protein S antigen 189 to 249 nmol/L inclusive (n = 15). The type I/III classification included family members with borderline total protein S antigen levels, possibly arising as a result of the overlap of antigen levels between normal and heterozygous individuals.26 This subgroup therefore removed potential complications from the genetic analysis that could have arisen from misclassification of individuals who displayed these borderline levels. However, for the purpose of statistical comparison, once genetic analysis was completed, the type I/III grouping was removed and family members were subclassified as either type I (n = 23) or type III (n = 8) protein S–deficient by strict adherence to the limits defined by the reference range. Anticoagulated protein S–deficient individuals (n = 13) were included in genetic investigations, but were excluded from detailed phenotypic analysis.
DNA sequencing of PROS1.The strategy used to examine the coding region and intron/exon boundaries of PROS1 has been described previously.34 In brief, oligonucleotide primers specific for PROS1 were used as primers in the polymerase chain reaction,35 and the amplification products were sequenced directly. Three individuals who were representative of the variable plasma phenotype were selected for genetic analysis. Individual VI-18 had a type I protein S–deficient phenotype (total protein S antigen, 175 nmol/L; free protein S antigen, 9 nmol/L; age, 15 years), V-34 had a type I/III phenotype (total protein S antigen, 207 nmol/L; free protein S antigen, 13 nmol/L; age, 38 years), and IV-30 had a type III protein S–deficient phenotype (total protein S antigen, 332 nmol/L; free protein S antigen, 6 nmol/L; age, 60 years). Individual IV-30 was the mother of V-34 and the grandmother of VI-18 (Fig 1).
All affected family members classified as type I or III protein S–deficient for genetic analysis (n = 17) were also investigated by direct sequencing for the presence of the Ser460Pro (protein S Heerlen) mutation,36 caused by a T to C transversion in exon XIII.
Screening of exon X by single-strand conformation polymorphism analysis.To screen all family members for the mutation identified in exon X, we used the Phastsystem (Pharmacia Uppsala, Sweden) to perform single-strand conformation polymorphism analysis (SSCP). The amplification product containing exon X of PROS1 was generated in essentially the same way as for sequencing. Equal quantities (3 μL) of amplification product and loading buffer (95% formamide, 20 mmol/L EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol FF ) were mixed and incubated at 95°C for 5 minutes to denature the double-stranded DNA, and were then chilled on ice. Pre-prepared homogenous 20% polyacrylamide gels and native buffer strips were used (Pharmacia). Gels were prerun for 100 aVh (250 V, 5 mA, and 2 W), at which time the samples were applied to the gel for 5 aVh. Electrophoresis continued for a total of 700 aVh, and DNA was visualized by a silver stain.37
Statistical analysis of phenotypic data.Laboratory data are expressed as the mean ± SD. The values were logarithmically transformed before analysis if required. Calculation of Pearson's correlation coefficient (r ) and probability (P ) and comparison by Student's t test were performed using Statview SE+ graphics software (Abacus Concepts Inc, Berkeley, CA). P less than .05 was considered significant in all cases.
RESULTS
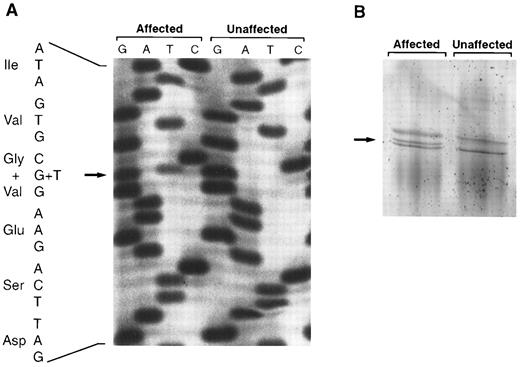
Genomic DNA amplification and sequencing of the entire coding region and intron/exon boundaries of PROS1, without interference from the pseudogene, was performed for the selected family members, individuals IV-30, V-34, and VI-18 (Fig 1). A single heterozygous mutation, Gly295Val (GGC to GTC) in exon X, was identified in all three individuals (Fig 2A). The mutant allele could be detected by SSCP, due to abnormal migration compared with normal homozygotes at this position (Fig 2B). Subsequent screening of 122 family members showed full cosegregation of the abnormal migration pattern and the mutation with reduced free protein S antigen levels, irrespective of plasma phenotype subclassification or anticoagulant treatment. All affected family members were heterozygous for the mutation. No other sequence abnormalities were identified in the three selected protein S–deficient individuals. In addition, none of 17 tested family members with type I or type III protein S deficiency had the protein S Heerlen allele.
(A) Identification of the Gly295Val mutation in exon X of PROS1 by single strand solid-phase sequencing. A section of the autoradiograph for an affected family member (reduced free protein S antigen level) and an unaffected relative (normal free protein S antigen level) is shown. The mutation was caused by a G to T transition (arrow). Specificity for PROS1 is evident, as the second base 3′ of the mutation is a guanine only; a difference between the PROS1 gene and the pseudogene has been reported at this position.20 (B) Detection of the Gly295Val mutation by SSCP. A fragment containing exon X of PROS1 was electrophoresed in a homogenous 20% polyacrylamide gel and stained with silver nitrate. The additional band seen in affected family members heterozygous for the Gly295Val mutation is indicated with an arrow.
(A) Identification of the Gly295Val mutation in exon X of PROS1 by single strand solid-phase sequencing. A section of the autoradiograph for an affected family member (reduced free protein S antigen level) and an unaffected relative (normal free protein S antigen level) is shown. The mutation was caused by a G to T transition (arrow). Specificity for PROS1 is evident, as the second base 3′ of the mutation is a guanine only; a difference between the PROS1 gene and the pseudogene has been reported at this position.20 (B) Detection of the Gly295Val mutation by SSCP. A fragment containing exon X of PROS1 was electrophoresed in a homogenous 20% polyacrylamide gel and stained with silver nitrate. The additional band seen in affected family members heterozygous for the Gly295Val mutation is indicated with an arrow.
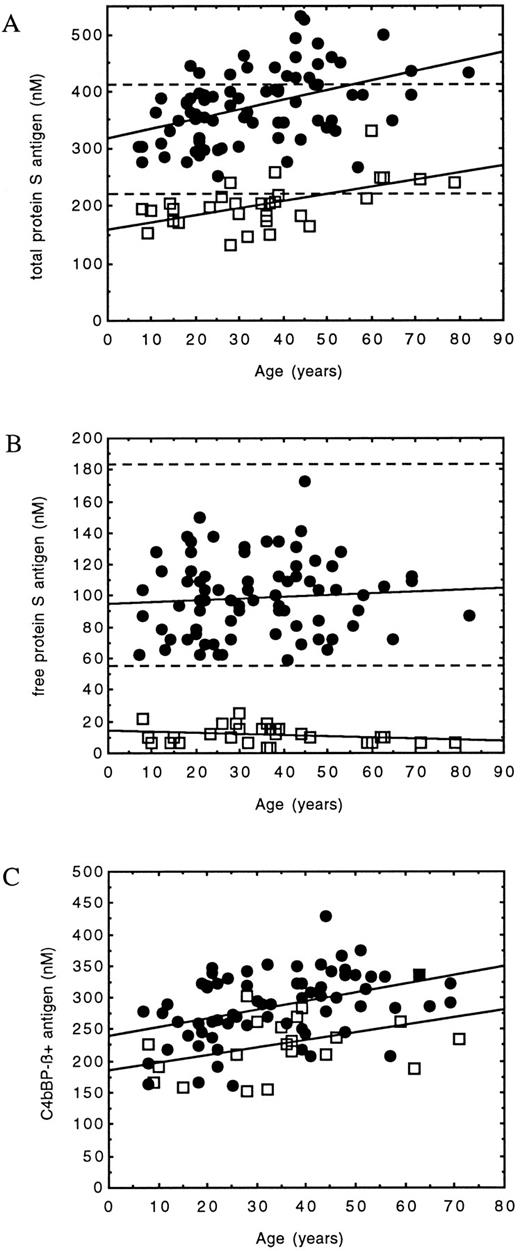
Taken overall, protein S–deficient family members with the Gly295Val mutation had significantly lower total protein S (203 ± 40 v 375 ± 63 nmol/L, P < .001), free protein S (11 ± 6 v 99 ± 24 nmol/L, P < .001), and C4bBP-β+ (228 ± 49 v 287 ± 54 nmol/L, P < .001) antigen levels than the unaffected relatives. Analysis of phenotypic data also showed evidence for an influence of age on total protein S and C4bBP-β+ antigen levels, but not on free protein S antigen. Significant correlations between total protein S antigen levels and age were defined for both protein S–deficient and normal family members (Table 1 and Fig 3A). No correlation was found in either group between free protein S antigen and age (Table 1 and Fig 3B). C4bBP-β+ antigen levels correlated significantly with age in normal family members, and although this did not reach significance in protein S–deficient family members (P = .053), there was a tendency for levels to increase (Table 1 and Fig 3C). C4bBP-β+ antigen was also found to be significantly correlated with total protein S antigen in both protein S–deficient (n = 21, r = .64, P < .01) and normal (n = 67, r = .68, P < .001) family members.
Graphic representation of the correlations between age and protein S–related determinants in normal (•) and protein S–deficient (□) family members (see Table 1). (A) Comparison between total protein S antigen and age. The limits of the reference range (219 to 407 nmol/L) have been superimposed (- - -). The increased level compared with the reference range as age increases can be clearly seen. Correlations were significant in both groups. Although this results in a type III protein S–deficient phenotype in some older individuals, the level is still reduced compared with that in normal family members of comparable age. (B) Comparison between free protein S antigen and age. The limits of the reference range (56 to 182 nmol/L) have been superimposed (- - -). In all protein S–deficient family members, the level remained well below the limit of the normal range. Neither of the correlations were significant. (C) Comparison between C4bBP-β+ antigen and age. The correlation was significant for normal family members, and although significance is not reached in protein S–deficient family members, there is a tendency for an increase.
Graphic representation of the correlations between age and protein S–related determinants in normal (•) and protein S–deficient (□) family members (see Table 1). (A) Comparison between total protein S antigen and age. The limits of the reference range (219 to 407 nmol/L) have been superimposed (- - -). The increased level compared with the reference range as age increases can be clearly seen. Correlations were significant in both groups. Although this results in a type III protein S–deficient phenotype in some older individuals, the level is still reduced compared with that in normal family members of comparable age. (B) Comparison between free protein S antigen and age. The limits of the reference range (56 to 182 nmol/L) have been superimposed (- - -). In all protein S–deficient family members, the level remained well below the limit of the normal range. Neither of the correlations were significant. (C) Comparison between C4bBP-β+ antigen and age. The correlation was significant for normal family members, and although significance is not reached in protein S–deficient family members, there is a tendency for an increase.
Formal analysis of protein S–deficient family members with a type III classification for phenotypic analysis (ie, total protein S antigen ≥219 nmol/L) showed that they were significantly older and had significantly higher total protein S and C4bBP-β+ antigen levels than those classified as type I protein S–deficient (Table 2).
Total protein S antigen levels are known to be influenced by a number of variables, including sex. Free and total protein S and C4bBP-β+ antigen were therefore examined with respect to sex for both normal and affected family members. Significantly lower free protein S antigen levels (91 ± 22 v 108 ± 24 nmol/L, P = .002) were found in unaffected female family members compared with males, but no significant difference was found in age or total protein S or C4bBP-β+ antigen levels. There was no significant difference for any of these variables in protein S–deficient family members (data not shown). Furthermore, regression analysis of data from unaffected male and female family members separately confirmed the independent influence of age on total protein S antigen levels. In both groups, a significant correlation was seen between these two variables (males: n = 38, r = .38, P = .02; females: n = 40, r = .56, P = .0002).
DISCUSSION
Of the well-established genetic risk factors for venous thromboembolism, protein S deficiency is perhaps the most difficult to diagnose with accuracy. Unlike antithrombin, protein C, and factor V, protein S can be complexed with a protein of the complement pathway, C4bBP-β+. Expression of protein S activity in plasma is determined in part by this binding interaction, with only the uncomplexed form of protein S available to act as a cofactor to APC. Consequently, there is uncertainty regarding which assay is best suited for diagnosis of protein S deficiency and is also predictive of risk for thromboembolism. Uncharacteristically, an effective diagnosis of protein S deficiency may not depend primarily on a measurement of total antigen, but only on the fraction that is available for one of its activities in plasma (ie, free protein S antigen). This diagnostic uncertainty has been highlighed by our recent finding of the coexistence of different protein S–deficient subtypes, type I and type III, in the same family. It can be noted that the distinction between these plasma phenotypes is on the basis of total protein S antigen levels, which are reduced in type I protein S deficiency alone.
We have investigated a large kindred that included individuals with both type I and type III protein S deficiency. Molecular genetic investigation showed a sole heterozygous mutation within the coding region and intron/exon boundaries of PROS1 in each of three subjects representative of the variable phenotype. The mutation would result in the amino acid substitution Gly295Val in the SHBG domain of protein S if the abnormal allele was expressed. This mutation cosegregated perfectly with diagnosis of phenotypic protein S deficiency (reduced free protein S antigen levels, subtypes I, III, I/III, and unclassified protein S–deficient individuals receiving oral anticoagulant treatment) within 122 members of the family group and has also been found to be associated with an increased risk of venous thromboembolism (Simmonds et al, unpublished observations, November 1996). The protein S Heerlen allele (allele frequency, ∼0.5%36) was considered a potential cause of type III protein S deficiency, as abnormal binding of the gene product to C4bBP has been demonstrated.38 However, it was absent from 17 type I or type III protein S–deficient family members. Furthermore, there was no possible influence of the factor V Arg506Gln mutation, since only one family member, with normal protein S antigen levels, was heterozygous at this locus.
Because all protein S–deficient family members were heterozygous for the same single mutation and no other genetic difference could be identified, the phenotypic data were examined for an explanation of the variable phenotype. We noted previously that individuals with a type III phenotype tended to be older,27 a finding that is also apparent in this family (Fig 1). When studied formally, the mean age in type III protein S–deficient relatives was found to be significantly higher than in those classified as type I protein S–deficient (Table 2). In addition, a significant correlation between total protein S antigen levels and age was determined in protein S–deficient family members. This resulted in an increase of total protein S levels to within the normal range or at borderline levels in older subjects (Fig 3A). There was no correlation between free protein S antigen levels and age (Fig 3B), which remained well below the lower limit of the reference range throughout the lifetime.
The increase in total protein S antigen levels with age was not limited to protein S–deficient family members, as they were also correlated in normal individuals. In this case, measurements showed a tendency to move above the upper limit of the reference range beyond 60 years (Fig 3A). Age was not associated with an increase in free protein S antigen (Fig 3B); however, the level always remained within the limits of the normal range.
The significant correlation defined between total protein S antigen and age in normal family members was maintained if male and female data were analyzed separately. It was not possible to establish a significant correlation between total protein S antigen and age in males and females separately within the protein S–deficient group, due to the low number of subjects (13 males and 18 females). However, there was no significant difference in either total protein S antigen or age between the two sexes among either normal or affected relatives. Interestingly, free protein S antigen levels were significantly lower in females compared with males within the normal group. However, levels always remained within the normal range. The physiologic relevance of this finding, also noted by other studies,17,39 is unknown, but has been attributed to hormonal influence. These data strongly suggest that age has an independent effect on the circulating concentration of total protein S antigen, an effect that is not subverted by the previously demonstrated effect of sex.17 39-43
Although total protein S antigen levels increased with age in both normal and affected groups, they still tended to be reduced in protein S–deficient family members compared with unaffected relatives of similar age (Fig 3A). This would be expected in a quantitative or type I protein S deficiency, almost certainly caused by the Gly295Val mutation identified, due to low stability of mRNA transcripts or production of protein unsuitable for secretion and subsequently sequestered in the Golgi apparatus. The amino acid residue affected is located within the SHBG domain of the protein S molecule and is conserved between protein S from different species, human SHBG, and rat androgen binding protein (data not shown). This supports a structural role for this residue and a potential cause of a type I phenotype upon alteration.
The amount of C4bBP-β+ antigen is an important factor in determining protein S phenotype, as it has been shown that the amount of free protein S antigen is equal to the molar excess of total protein S over C4bBP-β+.17 It was expected that C4bBP-β+ antigen levels in both normal and affected family members would increase with age, because they were significantly correlated with total protein S antigen levels in this family (P < .01), in agreement with previously published findings.27 This was confirmed (Fig 3C), although in the latter group the correlation did not reach significance (Table 1), probably due to the low number of data points (n = 21). However, C4bBP-β+ antigen levels were significantly higher in type III versus type I protein S–deficient individuals (Table 2). Therefore, the expected consequence of the relationships between total protein S, C4bBP-β+, and age would be stable free protein S antigen levels throughout the lifetime, as was observed.
Total protein S antigen levels have previously been shown to influence C4bBP-β+ antigen levels.27 As expected in this study, subjects (both protein S–deficient patients and a patient control group) receiving vitamin K–antagonist drugs (such as warfarin) had reduced protein S antigen levels, since it is a vitamin K–dependent protein. The levels of C4bBP-β+ antigen, which is not vitamin K–dependent and therefore would not be expected to be affected by warfarin, were also significantly reduced. This supports the contention that protein S antigen levels play a role in the regulation of C4bBP-β+. Also, individuals with the Gly295Val mutation in the family currently under investigation had significantly reduced C4bBP-β+ antigen levels compared with normal family members, in addition to reduced total protein S antigen. Since free protein S deficiency in this family was linked to the PROS1 locus located on chromosome 3 and the gene for C4bBP is located on chromosome 1, the mutation identified is unlikely to directly influence C4bBP-β+ expression at the molecular genetic level. Therefore, the increasing protein S antigen levels with age observed in this family cannot be explained by an increase in C4bBP-β+ antigen levels.
We have found that the difference between type I and type III protein S–deficient phenotypes is not a result of alternative genotypes, but is due to an increase in total protein S antigen levels as age increases. This was independent of the influence of sex, and an associated increase in C4bBP-β+ antigen levels resulted in free protein S deficiency throughout the lifetime. The reason for this age-related increase in protein S has not been examined. The effect of age on total protein S antigen levels in the general population is assumed to be the same as in the unaffected family members of this kindred, as suggested in a previous report.41 This would further complicate the diagnosis of protein S deficiency by assessment of total protein S antigen levels, since the provision of adequate control groups will be difficult to achieve. The assay for free protein S antigen differentiated normal from affected individuals regardless of the use of age-matched controls, and we therefore consider this the preferential variable to be determined in the diagnosis of protein S deficiency.
The metabolism of protein S is still poorly understood, despite rapid progress in the understanding of the molecular basis of deficiency. It is possible to envisage several acquired and genetic factors that could influence the relationship between free and total protein S and C4bBP-β+ antigen levels. An important known genetic factor of this type is protein S Heerlen,36,38 although other as yet unidentified mutations in the PROS1 coding region, in the 5′ and 3′ untranslated regions, or in the gene coding for C4bBP may also affect this relationship. It might therefore be premature to extrapolate the current finding of an “acquired” rather than a genetic explanation for the type III protein S–deficient phenotype in this family to many other cases of type III protein S deficiency. However, although such defects could be a direct cause of type III protein S deficiency, it seems unlikely that any such defect would directly cause a change in phenotype within a family from type I to type III. In one study of the normal population, total protein S antigen levels were found to increase with age.41 Furthermore, the tendency for type III protein S–deficient individuals to be older than those with a type I phenotype has also been observed in a group of 17 affected families who exhibit both phenotypes.27 Therefore, age-related changes in total protein S antigen could be a common mechanism for the coexistence of type I and type III protein S deficiency within the same kindred.
Supported by a Studentship and grants from the Special Trustees of Charing Cross Hospital and Medical School, a grant from the Swedish Medical Council (07143), grants from the Louis Jeantet Fondation de Medicine, the Österlund Trust, the King Gustaf V and Queen Victoria Trust, the Albert Påhlsson Trust, the Johan and Greta Kock Trust, and the Göran Gustavsson Trust, and research funds from University Hospital, Malmö, the Ann Lisa and Sven-Eric Lundgrens Trust for Medical Research, the Crafood Trust, Stiftelsen för blodsjukdomarsbekämpande, the Carin Trygger Trust (Swedish Medical Society), the Carl-Bertil Laurell Nordic Fund, and the Swedish Society for Medical Research.
Address reprint requests to Rachel E. Simmonds, BSc, Department of Haematology, Charing Cross and Westminster Medical School, St Dunstan's Road, Hammersmith, London, W6 8RP, UK.