Abstract
We examined human T-lymphotropic virus type I (HTLV-I) DNA integration in 68 patients with adult T-cell leukemia/lymphoma (ATL) by Southern blotting using EcoRI, which does not cut within the 9 kb of the genome and probes for pX and gag-pol region of HTLV-I. We detected defective proviral integration as a monoclonal band of various sizes with the pX but not with the gag-pol probe, or a monoclonal band of less than 9 kb with the pX probe, in 20 patients (29.4%). These were designated defective (D) type. With both probes, a single band greater than 9 kb was detected in 34 (50.0%), designated complete (C) type, and two or more bands greater than 9 kb, were designated multiple (M) type, in 14 (20.6%). Advanced age, a high LDH value, and hypercalcemia were more frequent in D type patients. The median survival time (MST) was 6.8, 24.4, and 33.3 months, for D, C, and M types, respectively (log rank P = .006). Among 52 sequentially examined patients, the HTLV-I integration patterns changed in 4 (7.5%). In three of these four, the rearrangements of the T-cell receptor (TCR)b gene concomitantly changed, suggesting the appearance of a new ATL clone. Another patient had the same rearrangement of the TCRb gene, indicating clonal evolution. The HTLV-I integration pattern changed at crisis from indolent to aggressive ATL in three patients. These findings suggested that the HTLV-I integration patterns have clinical implications in ATL pathophysiology. In contrast to the clonal evolution characteristic of the multistep carcinogenesis of most human malignancies, the frequent clonal change of ATL at crisis is a peculiar phenomenon, probably reflecting the emergence of multiple premalignant clones in viral leukemogenesis as suggested in Epstein-Barr virus associated lymphomagenesis in the immunocompromised host.
HUMAN T-LYMPHOTROPIC virus type I (HTLV-I) is etiologically associated with adult T cell leukemia/lymphoma (ATL), and the diverse clinical features of this disease have led to its subclassification; acute, lymphoma, chronic, and smoldering types.1-3 The median survival time (MST) is 6.2, 10.2, and 24.3 months for the first three types, respectively. An MST has not been established for the smoldering type.3 Advanced PS, high LDH level, age >40 years, large number of lesions, and hypercalcemia are the major factors associated with a shorter survival period in all subtypes of ATL according to a multivariate analysis.4 There are several other prognostic factors, such as ki67 positivity on ATL cells, immunophenotypes other than typical CD4+/CD8− ATL cells, and the thymidine kinase or soluble interleukin-2 (IL-2) receptor levels in serum.5-8
The incidence of ATL is rather low among HTLV-I carriers, and those who develop the disease usually have a 40- to 60-year latency period from the time of infection to progression to ATL.9 It has been postulated that the leukemogenesis of ATL consists of five steps, based on a stastical analysis of ATL development according to age.10 The tumor suppressor genes such as p53, p15, and p16, are abnormal in aggressive ATL.11 12 Sometimes, the abnormalities appear at the time of crisis. However, the precise mechanism of multi-step carcinogenesis in ATL remains unknown.
In all types of ATL, the monoclonal integration of the HTLV-I proviral DNA into tumor cells can be detected by Southern blotting.13 When the DNA of the tumor cells is digested with the endonuclease EcoRI, which does not cut within the 9 kb of the full HTLV-I genome, Southern blotting usually shows a single band of over 9 kb. However, sometimes there are patterns of multiple or defective HTLV-I integration.13,14 It is still controversial whether or not the integration pattern is of clinical significance.15-17 We describe here that the pattern of defective HTLV-I integration tends to be associated with aggressive ATL, and that the multiple (M), complete (C), and defective (D) types in that order, are apparently associated with prognosis from good to bad. Furthermore, we found a frequent clonal change in patients with ATL at the time of crisis.
PATIENTS AND METHODS
Patients.We assessed sixty-eight patients with ATL. The DNA analysis was sequentially analyzed in 52 of them. Peripheral blood (PB) and samples from other sites such as lymph nodes (LN) were examined at onset in 20 patients. These participants had been admitted and/or referred to our hospital since 1985. They were all Japanese and born in an area endemic for ATL, the Nagasaki district. The last follow-up date of this study was November 1995. The diagnosis of ATL was based on clinical features, immunophenotypes, anti–HTLV-I antibody, and the monoclonal integration of HTLV-I proviral DNA.18 The subgroups of ATL were defined as described.3
Anti–HTLV-I antibody and immunophenotypes.Serum samples were examined for antibody to HTLV-I by immunofluorescence assay and/or enzyme immuno-assay.19,20 Mononuclear cells (MNC) were separated from heparinized blood or LN cell suspension by Ficoll-Conray density gradient centrifugation. The surface phenotypes of ATL cells were determined with a panel of monoclonal antibodies using an immunofluorescence assay or a Spectrum III apparatus (Ortho Diagnostic Systems, Raritan, NJ) as described.6
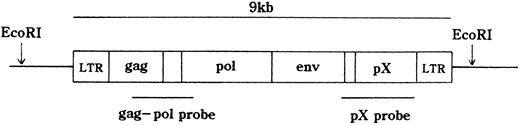
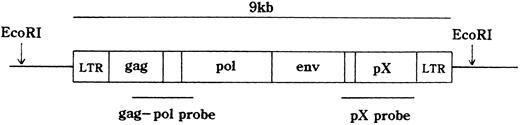
HTLV-I proviral DNA integration and T-cell receptor (TCR)b gene rearrangement.The integration of the HTLV-I provirus genome was assayed as described.18 21 In brief, high molecular weight DNA was extracted from MNCs. DNA samples (10 μg) were digested with restriction enzymes and size fractionated on 0.7% agarose gels. They were then electro-transferred onto a nitrocellulose membrane and hybridized to randomly primed 32P-labeled DNA probes specific for HTLV-I or TCRb genes. Thereafter, the blots were washed at appropriate stringency and visualized by autoradiography. EcoRI, which does not cut within the HTLV-I provirus genome was used for HTLV-I. EcoRI, HindIII, or BamHI was applied to TCRb. The probes were a 2.3-kb BamHI, Sma I fragment of the pX region, a 1.6-kb Pst I fragment of the gag-pol region of HTLV-I gene (Fig 1), and a 0.9-kb Pst I fragment of the TCR Cb2 chain gene.
Statistics.Survival was calculated according to the Kaplan-Meier method and compared by means of the log rank test. The incidence of clinical features among the subgroups was analyzed by means of Pearson's Chi-square test or the Mann-Whitney U-test.
RESULTS
Southern blots of the HTLV-I provirus and TCRb gene.Using EcoRI, which does not cut within the 9 kb of the HTLV-I genome, Southern blots both for pX and gag-pol probes revealed a single band of >9 kb in 34 of the 68 patients (50%) with ATL at onset (Fig 2, Table 1). These patients were considered to have a single complete HTLV-I genome and were designated C type. Fourteen patients (20.6%) showed two or more bands >9 kb with both pX and gag-pol probes (10 with 2 bands, and 4 with 3 bands). These 14 patients were considered to have multiple copies of the complete HTLV-I genome and were designated as M type. Seven patients (10.3%), who showed at least one band <9 kb with the pX probe, were considered to have an integrated defective provirus and were designated D type. In six of these seven patients, the band <9 kb was not detected with the gag-pol probe. The other 13 patients (19.1%) showed one or more bands of >9 kb with pX probe, but one of the band(s) was not detected with the gag-pol probe. These 13 patients were also considered as D type. Four of the 20 D type patients showed two bands with the pX probe and one band with the gag-pol probe, meaning that one HTLV-I provirus was D type, whereas the other was probably C type. Monoclonality was also confirmed in all patients by TCRb gene rearrangement (Fig 2). Restriction enzyme digestion produced one or two, but never more than two rearranged bands even when there were three integrated HTLV-I bands. This finding, in addition to the detection of multiple HTLV-I integration on different chromosomes by in situ hybridyzation and similar phenomena in bovine leukemia virus-related leukemia, suggested that the M type represents multiple copies of the HTLV-I provirus per cell.22 23
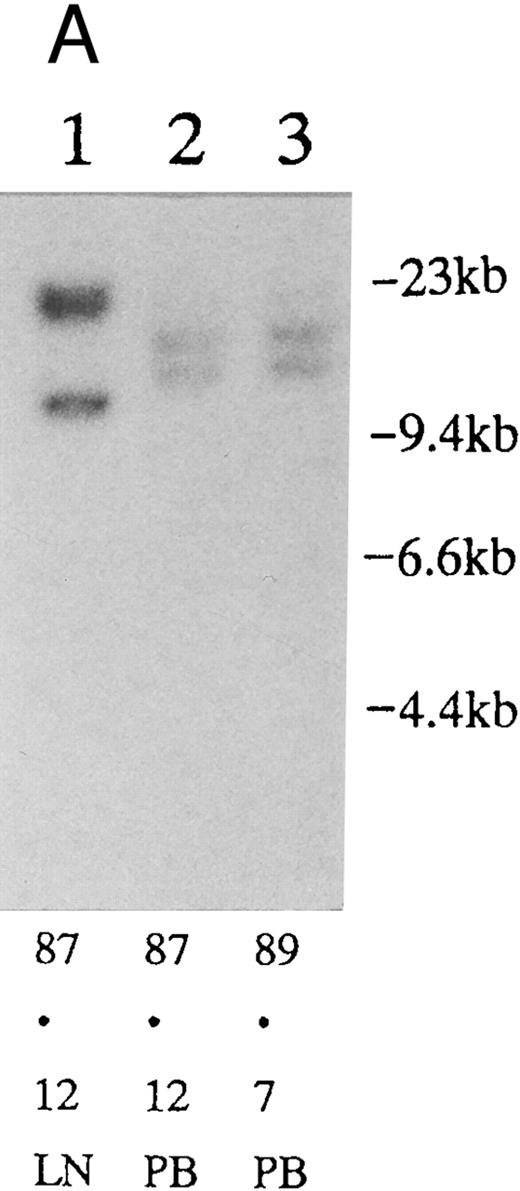
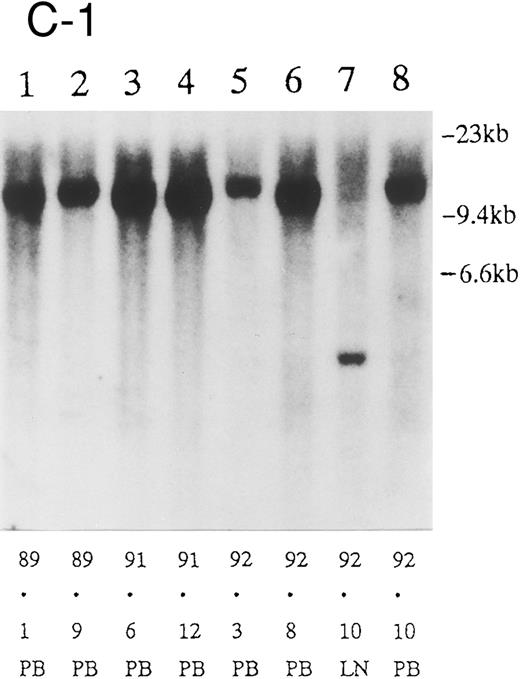
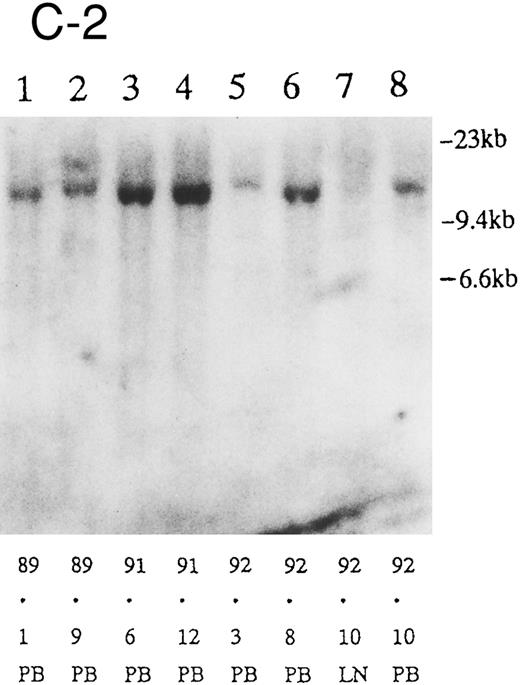
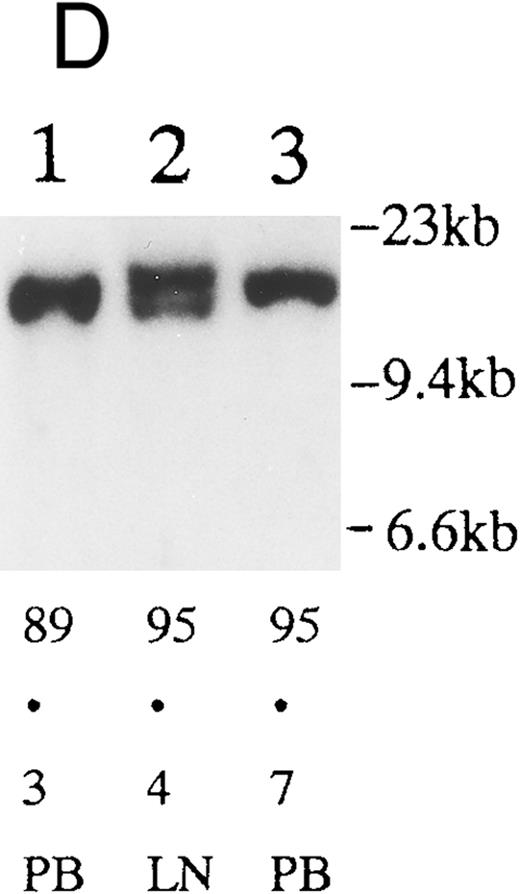
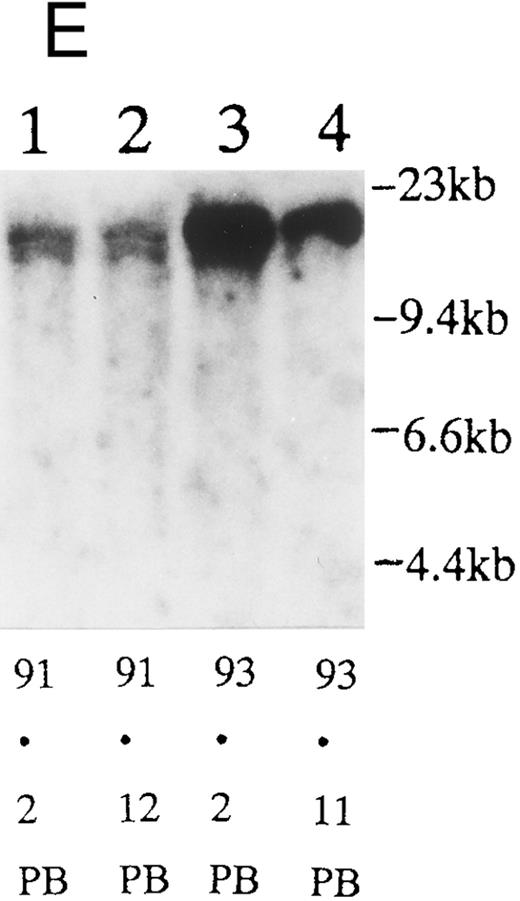
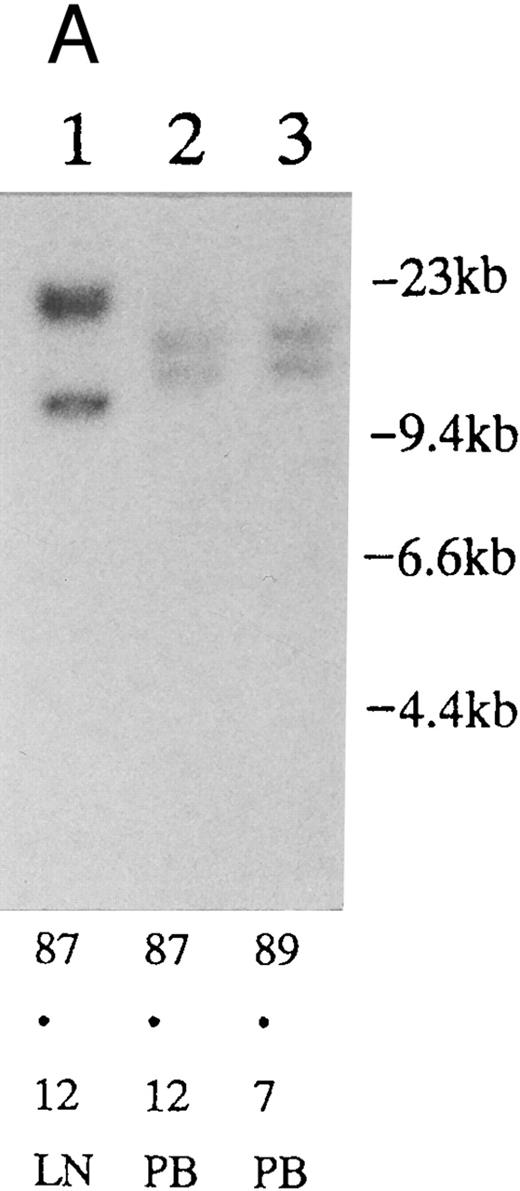
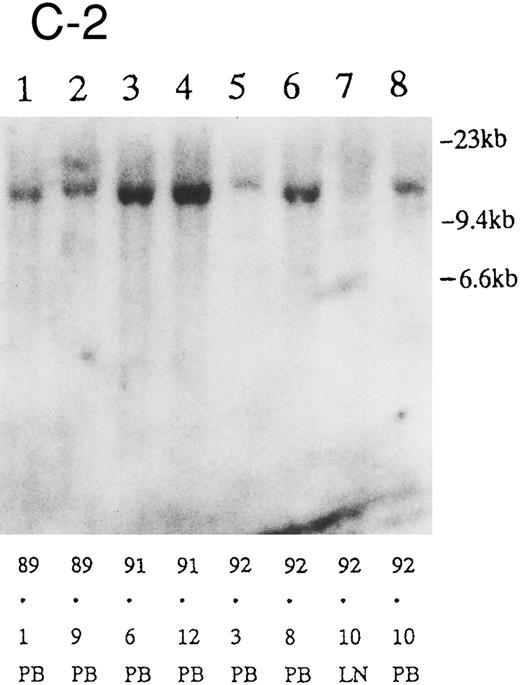
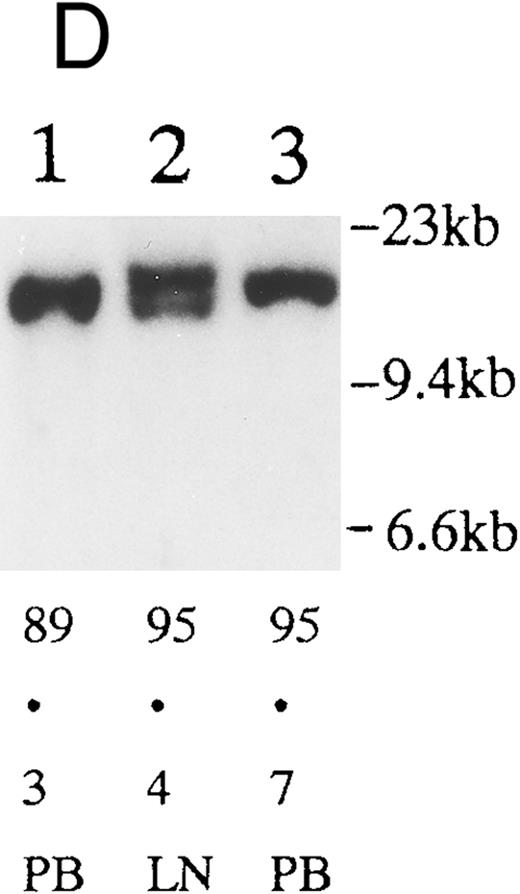
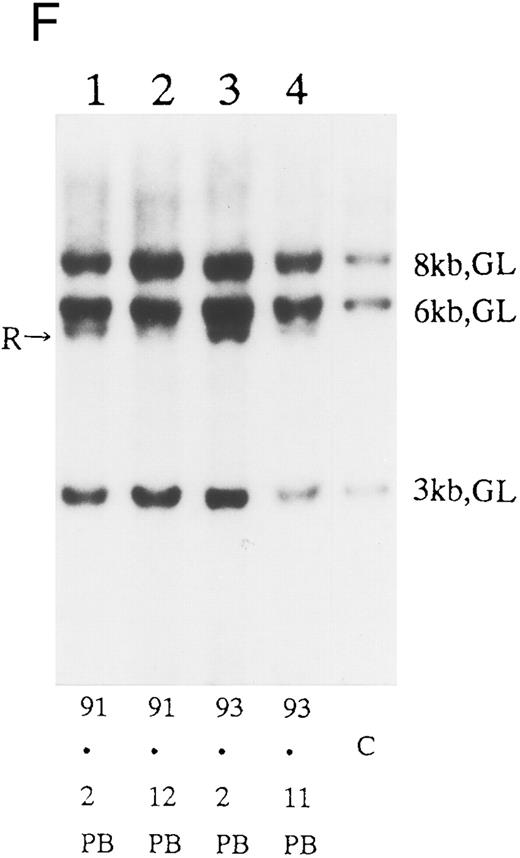
(A) Sequential analysis of proviral HTLV-I integration in patient 1. Cellular DNA of MNC was digested by EcoRI and hybridized with the pX probe. Two bands larger than 9 kb, (M type), were detected in LN at onset (lane 1), while same sized M type bands, distinct from those in lane 1, were detected both in PB at onset (lane 2) and in PB at relapse (lane 3). (B) Sequential analysis of TCRb gene rearrangement with HindIII in patient 1. (C) Control DNA from HL-60. Samples in lanes 1 to 3 represent those in lanes 1 to 3 in (A). Lane 1, one rearranged band, lanes 2 and 3, two rearranged bands of the same size that were distinct from the rearranged band in lane 1 (GL, germ line; R, rearranged band). (C-1) Sequential analysis of proviral HTLV-I integration in patient 2. One band >9 kb, (C type) was detected in all PB during the course with the pX probe, while one band <9 kb, (D type) was detected in LN at crisis (lane 7). (C-2) Samples in lanes 1 to 8 represent those in lanes 1 to 8 in (C-1). With the gag-pol probe, the same size of C type band as that shown in (C-1) was detected in lane 1 to 6 and 8, while the D type band with the pX probe was not detected with the gag-pol probe in lane 7. (D) Sequential analysis of proviral HTLV-I integration in patient 3. The same C type band was detected in PB at onset and at remission (lanes 1 and 3) with the pX probe. M type bands, of a size distinct from the C type, were detected in LN at crisis (lane 2). (E) Sequential analysis of proviral HTLV-I integration in patient 4. The same size of M type bands was detected at onset (lane 1) and just before chemotherapy (lane 2) in PB with the pX probe. The density of one of the two bands decreased and that of the second increased at crisis (lane 3), whereas the second was only detected at the terminal stage (lane 4). (F) Sequential analysis of TCRb rearrangement with HindIII in patient 4. Samples in lanes 1 to 4 represent those in lanes 1 to 4 in (E). One rearranged band of the same size was detected in lanes 1 to 4.
(A) Sequential analysis of proviral HTLV-I integration in patient 1. Cellular DNA of MNC was digested by EcoRI and hybridized with the pX probe. Two bands larger than 9 kb, (M type), were detected in LN at onset (lane 1), while same sized M type bands, distinct from those in lane 1, were detected both in PB at onset (lane 2) and in PB at relapse (lane 3). (B) Sequential analysis of TCRb gene rearrangement with HindIII in patient 1. (C) Control DNA from HL-60. Samples in lanes 1 to 3 represent those in lanes 1 to 3 in (A). Lane 1, one rearranged band, lanes 2 and 3, two rearranged bands of the same size that were distinct from the rearranged band in lane 1 (GL, germ line; R, rearranged band). (C-1) Sequential analysis of proviral HTLV-I integration in patient 2. One band >9 kb, (C type) was detected in all PB during the course with the pX probe, while one band <9 kb, (D type) was detected in LN at crisis (lane 7). (C-2) Samples in lanes 1 to 8 represent those in lanes 1 to 8 in (C-1). With the gag-pol probe, the same size of C type band as that shown in (C-1) was detected in lane 1 to 6 and 8, while the D type band with the pX probe was not detected with the gag-pol probe in lane 7. (D) Sequential analysis of proviral HTLV-I integration in patient 3. The same C type band was detected in PB at onset and at remission (lanes 1 and 3) with the pX probe. M type bands, of a size distinct from the C type, were detected in LN at crisis (lane 2). (E) Sequential analysis of proviral HTLV-I integration in patient 4. The same size of M type bands was detected at onset (lane 1) and just before chemotherapy (lane 2) in PB with the pX probe. The density of one of the two bands decreased and that of the second increased at crisis (lane 3), whereas the second was only detected at the terminal stage (lane 4). (F) Sequential analysis of TCRb rearrangement with HindIII in patient 4. Samples in lanes 1 to 4 represent those in lanes 1 to 4 in (E). One rearranged band of the same size was detected in lanes 1 to 4.
Clinical features according to integration pattern.As shown in Table 1, in aggressive ATL (41 acute type and 6 lymphoma type) the incidences of the C, M, and D types were 44.7%, 19.1%, and 36.2%, respectively, while in indolent ATL (19 chronic type and 2 smoldering type) they were 61.9%, 23.8%, and 14.3%, respectively. The prognostic factors showed that D type patients more frequently had poor prognostic factors, such as advanced age, a high LDH value, or hypercalcemia than the C or M type patients. Among 60 samples immunologically examined, nine had a CD4 and/or CD8 antigen abnormality as shown in Table 1. D type patients had this unusual phenotype more frequently (27.8%) than the other two types (C type, 6.3%; M type, 20%), although the differences were not statistically significant.
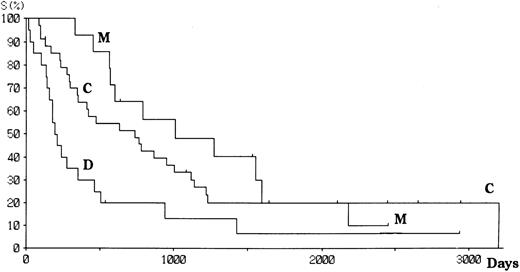
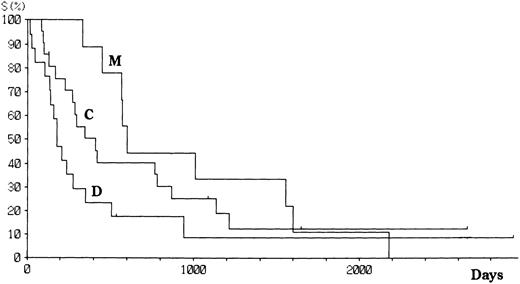
Patients with aggressive ATL were treated with doxorubicin, vincristine, cyclophosphamide, prednisolone, and one or several other antineoplastics.18 Patients with indolent ATL were followed up with no or mild chemotherapy (eg, prednisolone or cyclophosphamide alone) until the disease progressed. Fifty-six patients have died since the study started, and 12 remain alive with or without disease. The projected actual survival curves for each type of HTLV-I integration are shown in Fig 3. At a median follow-up time of 18.5 months, the median survival time (MST) was 33.3, 24.4, and 6.8 months for M, C, and D type patients, respectively (log rank P = .006). However, there was no difference in the number of long-term survivors between the three groups. Among aggressive ATL, the MST were 19.7, 9.8, and 4.6 months, respectively, for M, C, and D types. The survival of patients with aggressive ATL was similar among all subtypes (Fig 4), although the significance of the survival difference disappeared.
Survival of patients with ATL according to the integration pattern of HTLV-I provirus.
Survival of patients with ATL according to the integration pattern of HTLV-I provirus.
Survival of patients with aggressive ATL according to the integration pattern of HTLV-I provirus.
Survival of patients with aggressive ATL according to the integration pattern of HTLV-I provirus.
Clonal analysis in different sites and sequential analysis.PB and other samples (13 lymph node, 3 pleural effusion, 1 ascites, 2 broncho-alveolar lavage fluid, 1 cardiac tumor) from 20 patients were examined by Southern blotting at onset. Nineteen patients had the same size of HTLV-I integration bands in both lesions, indicating the same clone at different sites. However, in one patient, we detected M-type integration of HTLV-I proviral DNA at different sites of the genomic DNA in PB and LN cells (Fig 2A and Table 3; patient 1). Furthermore, samples from this also showed different rearrangements of the TCRb gene (Fig 2B). With intensive chemotherapy this patient achieved partial remission (PR) with the disappearance of LN swelling and normalization of the LDH value. However, a low ratio of ATL cells was detected even at remission in the PB. After approximately 19 months of remission, a lesion appeared in the spine. Even after relapse, the morphologic, immunologic, and molecular findings of PB remained stable (Fig 2, Tables 2 and 3).
We sequentially examined the HTLV-I DNA integration pattern of 52 ATL patients. The median number of the examination was 3 (range, 2-9) and the median follow-up duration was 16 months (range, 2-49). The disease status ranged from indolent to indolent in 5, from indolent to crisis in 12, relapse after complete remission (CR) of aggressive ATL in 5, relapse after PR of aggressive ATL in 20, and no remission of aggressive ATL in 10. Three (25%) of the 12 patients who underwent crisis from indolent ATL showed a changed HTLV-I DNA integration pattern (Fig 2C, D, and E). The other 49 patients, including three with a change in the immunophenotypes of ATL cells at the time of exacerbation, showed the same HTLV-I bands during the course. The characteristics of the three patients with a change of HTLV-I integration at the time of crisis and patient 1 described above are summarized in Tables 2 and 3. The changes in HTLV-I integration were from C to D type, from C to M type, and from M to C type, respectively, in patients 2, 3, and 4 (Table 3, Fig 2). In patient 4, one of the two HTLV-I integration bands was detected at the same position during the course, whereas the other disappeared after crisis. This patient showed the same rearranged bands of TCRb during the course of the disease (Fig 2F). In this patient, the ATL clone underwent clonal evolution. The morphology of the ATL cells in PB changed from mature to blastic at the time of clonal evolution. In the other three patients (1, 2, and 3) with a different HTLV-I integration pattern, the rearranged patterns of TCRb genes also differed (Fig 2B for patient 1; data not shown for patients 2 and 3). These findings suggested the appearance of a new ATL clone distinct from that at onset, because the integration sites of HTLV-I provirus are random.24 In patient 2, who underwent a clonal change from C type in PB at onset to D type in LN at crisis, the phenotype of ATL cells changed from CD4+, CD8− in PB to CD4+, CD8+ in LN. After crisis, the disease rapidly progressed in this patient despite chemotherapy. On the other hand, in patient 3 who underwent a clonal change from the C type in PB to M type in LN without a phenotypic change, chemotherapy after crisis was successful. She is now in PR with no lymphadenopathy, but with a low ratio of ATL cells in PB, with an HTLV-I integration band the same size as that in PB at the chronic phase (Fig 2D, Tables 2 and 3).
DISCUSSION
Detection of monoclonal integration of HTLV-I proviral DNA by Southern blotting is particularly important in the diagnosis of ATL.13 We first used the pX probe for ATL diagnosis because a deletion of this region of HTLV-I has never been found in fresh ATL cells or in HTLV-I–integrated cell lines.14,15,18,21 We also used the gag-pol probe to assess with more accuracy the proportion of samples with 5′ deleted proviruses. The endonuclease EcoRI divides the HTLV-I integration pattern into C, M, and D types.13-16 The reported frequency of unusual integration patterns such as M and D types of HTLV-I provirus is variable. Konishi et al14 have reported the M type in 10 and D type in 3 of 48 patients with ATL. Shimamoto et al16 have reported the M type in 3 and the D type in 3 of 89 patients. They considered two or more bands as the M type, even when one of the two bands was smaller than 9 kb, whereas we considered them as the D type. According to our definition, the frequency of M and D types are 9 and 4 of 48 patients in Konishi's study, 0 and 6 of 89 in that of Shimamoto, 13 and 28 of 95 in that of Ohshima, and 14 and 20 of 68 patients in our study.14-16 Thus, the unusual integration pattern is rather frequent in ATL.
We first examined the clinical importance of the integration pattern of HTLV-I proviral DNA in patients with ATL. The survival of D type patients was significantlly worse than that of the M and D type patients, although there was no difference in the number of long-term survivors between the three groups. D type HTLV-I integration was frequently detected in aggressive ATL. Even in aggressive ATL, the survival trend differed among HTLV-I integration patterns. As mentioned above, four of the D type patients had multiple bands. Including them in the M type did not change the trend in the survival difference (data not shown). There has been a controversy over whether the integration pattern of the HTLV-I provirus has pathophysiologic significance. Ohshima et al have reported that the integration pattern does not correlate with the clinical subtypes of ATL, although the definition of the subtype and the survival of the patients were not described.15 Shimamoto et al16 have reported that patients whose samples had two bands after EcoRI digestion showed a worse clinical course, whereas those with one band smaller than 9 kb showed a more favorable clinical course than C type patients. On the other hand, D'Incan et al17 have described a patient in whom samples had three HTLV-I bands after EcoRI digestion and who showed only cutaneous lesions and had a good prognosis. According to our criteria, the three patients described by Shimamoto belong to the D type (two had one band >9 kb and one <9 kb; the other had two bands <9 kb), whereas the patient described by D'Incan et al was of the M type (3 bands >9 kb). It remains unknown why the three patients described by Shimamoto with one band <9 kb (defective HTLV-I) had a good prognosis, whereas most of the other D types, including our patients, had a poor prognosis. In our study, unusual immunologic phenotypes of ATL cells were frequent among the D type. Two of the patients with D type described elsewhere, showed an unusual phenotype, BM infiltration with giant and blastic ATL cells, and a rapidly progressive course.25 Because we and others analyzed the integration patterns only by means of Southern blotting, minimal defective virus, such as HTLV-I with defects in the LTR region, may be included in the C or M types. The polymerase chain reaction (PCR) has shown that at least 34% of patients with ATL carry deleted viral genome in their leukemic cells.26 Further studies with more sensitive methods will provide more accurate information from which to draw a conclusion concerning the clinical features in relation to the HTLV-I integration pattern.
Several oncogenes and tumor-suppressor genes are crucial in the multi-step carcinogenesis of human malignancies.27 For example, clonal evolution in which the rearrangement pattern of one of the two immunoglobulin heavy chain (IgH) gene changes and that of the other remains the same, is frequent in the exacerbating precursor, B-acute lymphoblastic leukemia.28 We sequentially examined HTLV-I integration during the course of ATL in 52 patients. Both the HTLV-I integration and TCRb rearrangement patterns changed in two of 12 patients in crisis from indolent ATL to aggressive ATL. This phenomenon suggested that a new ATL clone distinct from that at onset emerges at crisis. The new clone was detected only in LN in both patients. Furthermore, a patient with acute ATL (patient 1) had two distinct clones at onset, one in PB and the other in LN. This probably means that this patient also represents a crisis from smoldering ATL, because a residual ATL clone was detected only in PB during the remission phase after chemotherapy. On the other hand, clonal evolution was found only in one patient in crisis from smoldering ATL (patient 4). Both clones were detected in PB. In this patient, one of the two HTLV-I integration bands at onset disappeared after crisis, while the rearranged bands of TCRb were the same at both times. Although chromosomes were not examined, it is likely that chromosomal deletion occurred at clonal evolution in this patient. The other 48 patients showed the same HTLV-I integration and TCRb rearrangement during the clinical course irrespective of sites and relapse after remission with or without phenotypic changes. There are three reports of biclonal ATL.29-31 The first patient had a different clone in the PB at onset and at relapse after the spontaneous regression of ATL.29 The second had two different clones, one in PB and the other in LN at onset.30 In this patient, the ATL cells were CD4+, CD8− in PB and CD4+, CD8+ in LN. The third also had two different clones, one in CD4+CD8− cells in PB and the other in CD4−CD8+ cells in PB.31 In this patient, the CD4−, CD8+ cells expanded during disease exacerbation. In two of the three biclonal patients in our study, both of the ATL clones in each were CD4+, CD8−. The prognosis of patients with biclonal ATL in our study was diverse. Patients 1 and 3, who were M type at crisis in LN, achieved remission, whereas patient 2, who was D type with a phenotypic change at crisis in LN, had a rapidly proggressive disease. The integration patterns of HTLV-I seemed to have prognostic value even after a clonal change of ATL. Perhaps more biclonal ATL could be identified by extensive sequential Southern blotting for the HTLV-I and TCRb genes during the clinical course.
In contrast to the clonal evolution generally seen in the multi-step carcinogenesis of most human malignancies, a clonal change of ATL at the time of crisis, corresponding to the late stage of multi-step carcinogenesis, is unusual. It resembles the multi-clonal state associated with Epstein Barr (EB) virus-associated lymphoma detected by rearrangements of the IgH gene in an immunocompromised host.32,33 Two notions could explain the development of a clonal change in ATL. One is the oligo-clonal expansion of HTLV-I infected premalignant cells even in asymptomatic carriers, detected by inverse PCR or by linker ligation PCR.34,35 These oligo-clones in HTLV-I carriers may be the origin of a new ATL clone during crisis. The other is an immunosuppressive state documented in patients even in the early stage of ATL, or in HTLV-I carriers.36,37 There are three reports of ATL developing during immunosuppressive treatment in a renal transplant recipient.38-40 Many of the oligo-clonal EB virus-associated lymphomas in immunosuppressed individuals such as organ allograft recipients are reversed when immunosuppressive drugs are withdrawn.32 Thus, virus-induced leukemogenesis seems to have a unique oligo-clonal state distinct from the clonal evolution generally associated with human de novo malignancies.
The prognosis of patients with aggressive ATL is extremely poor. There is no curative therapy even for patients with indolent ATL. Thus, the prevention or early eradication of aggressive crisis from indolent ATL or de novo aggressive ATL from the carrier state of HTLV-I is of clinical importance. We reported that HTLV-I carriers with the monoclonal proliferation of T lymphocytes have a far better prognosis than patients with chronic ATL.21 Early detection of either a clonal change or clonal evolution may provide a new therapeutic approach.
In conclusion, the integration patterns of the HTLV-I provirus have clinical implications for ATL. The multiple, complete and defective types in that order are apparently associated with prognosis from good to bad although there were no differences in the number of long-term survivors. ATL frequently underwent a clonal change at the time of crisis.
Supported in part by a Grant-in-Aid for Cancer Research (2S-1) from the Ministry of Health and Welfare of Japan.
Address reprint requests to Kunihiro Tsukasaki, MD, Department of Hematology, Atomic Disease Institute, Nagasaki University School of Medicine, Sakamoto 1-12-4, Nagasaki 852, Japan.