Abstract
Fetal cells that circulate in maternal peripheral blood (PB) during pregnancy offer a potential source of nucleated fetal material for noninvasive prenatal diagnosis. Fluorescence-activated cell sorting was used to target two populations of fetal cells: nucleated erythroid cells (NECs; CD71/glycophorin-A+ CD45lo-int CD34−) and hematopoietic progenitor cells (CD34+ cells; CD34++ CD71/glycophorin-A− CD45int). Fetal cells were detected by fluorescence in situ hybridization (FISH) using directly conjugated chromosome X and Y probes in 65% (13 of 20) of the maternal PBs (fetal karyotype 46,XY). The frequency of fetal cells isolated from the NEC and CD34+ fractions was, respectively, 0 to 14 and 0 to 7 cells per 2 × 107 previously frozen maternal cells (≈20 mL of blood). In nonfrozen samples, the yield and recovery of fetal cells was moderately improved. Culturing the CD34+ sorted fractions in serum-free media with cytokines improved the quality of the FISH preparations and resulted in a slight expansion in detectable fetal cells. The frequency of fetal cells isolated from cultured CD34+ fractions was 0 to 35 and 0 to 93 cells per 2 × 107 previously frozen and nonfrozen maternal PB cells, respectively. These results document the isolation, characterization, and enumeration of fetal cells from the maternal periphery that appear to be present in most, but not all, samples analyzed.
CURRENTLY, PRENATAL DIAGNOSIS of fetal aneuploidies involves either chorionic villi sampling or amniocentesis, both invasive procedures that carry a risk, primarily to the fetus. The isolation of fetal cells from the maternal circulation has the potential to allow early noninvasive genetic analysis without endangering the fetus.1-3 However, targeting these cells has proved to be technically challenging because of their low frequency and the absence of suitable fetal cell markers and corresponding monoclonal antibodies (MoAbs). To be useful in prenatal diagnosis, fetal cells must be distinguished from the vast majority of maternal cells, they must be enriched to an acceptable level of purity, and they must be unequivocally identified as cells of fetal origin.
The reported frequencies of fetal cells in normal maternal peripheral blood (PB) varies widely, ranging from 1 in 5,000 to 1 in 108 nucleated cells.4-6 Factors that may influence these frequencies include the type of fetal cell analyzed, the gestational age at the time of sampling, and the accuracy of methods to enrich, identify, and quantify the fetal cell target population. The incidence of fetal cells in maternal blood has been reported to increase with advancing gestation5 and after toxemia, abortion, cesarean section,7 amniocentesis,8 or bimanual pelvic examination9 and in pregnancies in which the fetal and placental karyotype was abnormal.10,11 In view of the spurious or negative results of some investigations, it has become clear that specific and sensitive techniques with appropriate controls to enrich for fetal cells and confirm their fetal origin are essential, a requirement not always met in previous studies.8,12 13
Despite some promising results and several investigations in this field, many of the basic questions regarding the circulation of fetal cells in the maternal blood remain unanswered. Specifically, more information on the frequency and type of fetal cells that circulate during pregnancy needs to be established, the possibility of clonally expanding fetal cells needs to be explored, and techniques that will facilitate the identification and isolation of fetal cells need to be developed before this technique can be implemented for routine prenatal screening. In this study we aimed to measure the frequency of nucleated erythroid cells (NECs)14-17 and CD34+ progenitor cells18-20 of fetal origin in maternal blood. Several studies have focused on NECs because (1) they are present in the maternal circulation as early as the first trimester, (2) they are mononucleated committed progenitor cells with limited, if any, proliferative potential and a relatively short half-life, and (3) these cells are rare, if not absent, in the normal adult periphery.4,21,22 CD34+ cells were targeted because rare fetal progenitors cells could perhaps be clonally expanded, therefore allowing more extensive or reliable genetic analysis. The presence of CD34+ fetal cells in maternal blood has been documented.18-20 However, the observation that CD34+ fetal cells may include cells from past pregnancies has tempered enthusiasm for strategies targeting these cells for prenatal diagnostic purposes.
In the present study we used combination of MoAbs specific for CD71, glycophorin-A, CD45, and CD34 to enrich either NECs or CD34+ progenitor cells. Sorted cells were analyzed by fluorescence in situ hybridization (FISH) to detect and quantify male cells of presumed fetal origin. Previously frozen or nonfrozen maternal PB specimens from normal and abnormal pregnancies (fetal karyotype; trisomy 18 and 21) and PB samples from women who had never been pregnant (nongravida) were analyzed without prior knowledge of the fetal karyotype or donor origin. Samples from pregnant women were obtained at 10 to 13 weeks' gestation, before any invasive prenatal diagnostic procedure.
MATERIALS AND METHODS
Cells.PB samples from male and female (gravida and nongravida) donors were obtained with informed consent and according to institutional guidelines approved by the Clinical Screening Committee for Research Involving Human Subjects of the University of British Columbia (Vancouver, BC, Canada). Approximately 20 mL of maternal PB was drawn from pregnant women before any invasive prenatal diagnostic procedure at 10 to 13 weeks' gestation. Information regarding the past and present obstetrical history as well as results of fetal karyotyping obtained by conventional methods (amniocentesis and chorionic villi sampling) was obtained with the consent of the gravidae. Samples were processed without previous knowledge of fetal karyotype or donor gravid status to avoid sampling bias. Low-density cells (<1.077 g/mL) from male and female donors were isolated using Ficoll-Paque (Pharmacia LKB, Uppsala, Sweden), washed twice in phosphate-buffered saline (PBS) and resuspended in Hanks' HEPES-buffered salt solution containing 50% fetal calf serum (FCS) and 7.5% dimethylsulfoxide (DMSO), aliquoted, and frozen in replicate until used. A maternal PB cryo bank consisting of 200 maternal PB specimens and information regarding gestational age, obstetrical history, and fetal karyotype information was established for use in this study. Frozen control PB samples consisting of artificial mixtures of nongravida female and male mononuclear cells at a ratio of 98:2 were used to establish uniform flow cytometry sort parameters and provide controls for FISH analysis. Vials of frozen control and maternal PB were rapidly thawed and slowly diluted with Dulbecco's modified eagle medium (DMEM; Stem Cell Technologies, Vancouver, BC, Canada) containing 30% FCS and 0.1 mg/mL DNA-se (type II-S, D4513; Sigma Chemical Co, St Louis, MO). Cells were then washed twice and resuspended in Hanks' HEPES-buffered salt solution containing 2% FCS and 0.1% sodium azide (HFN) plus 5% normal human serum to block Fc receptors and for subsequent staining.
Red blood cells (RBCs) preserved in Alsever's solution [1:1 ratio of RBCs to Alsever's; dextrose (114 mmol/L):trisodium citrate, dihydrate (27 mmol/L):sodium chloride (72 mmol/L)] for up to 6 weeks from a nongravida female donor were added to the thawed control PB sample to a final concentration of 1 × 105 RBCs /1 × 107 nucleated cells. These RBCs were used to standardize flow cytometry settings. Fetal liver (FL; 10 to 16 weeks' gestation) samples were isolated by mechanical dispersion and flushing with DMEM supplemented with 10% FCS and were used as control cells for subsequent staining, flow cytometry, and FISH. In a separate set of experiments, 10 maternal PB samples were processed within 14 to 18 hours after venipuncture collection and without cryopreservation.
Labeling of cells and sorting.PB cells were stained as previously described.23 Briefly, cells (107/mL) were incubated simultaneously with MoAbs specific for CD34 (8G12 labeled with Cy5), CD71 (1C5 labeled with phycoerythrin [PE]), glycophorin-A (10F7-MN labeled with PE), and CD45 (9.4 labeled with fluorescein isothiocyanate [FITC]) for 30 minutes at 4°C. Controls for staining consisted of single-stained suspensions and cells stained with matched isotype control antibodies. Cells were then washed once with HFN and once with HFN containing 1 μg/mL propidium iodide (PI) and resuspended in HFN and 0.1 mg/mL DNA-se before sorting.
Flow cytometric analysis and cell sorting were performed with a FACStar Plus (Becton Dickinson, Mountain View, CA) equipped with argon and helium neon lasers. Ten-micron fluorescent beads were used to align the lasers and standardize the forward light scatter (FLS) threshold before each experiment. The flow cytometer was flushed with 10% household bleach for 5 minutes between samples to prevent cross-contamination of cells between samples. Specific fluorescence of FITC, PE, PI, and CY5 excited at 488 nm (0.4 W) and 633 (30 mW) as well as FLS and orthogonal light scatter (SSC) signals, were used to establish sort windows. PI was used to exclude dead cells. Positive events were defined as fluorescence that exceeded 99% of the events observed with matched-isotype control antibody conjugates. Two cell populations were targeted and simultaneously sorted: (1) NECs (CD71/glycophorin-A+ CD45lo-int CD34lo), and (2) hematopoietic progenitor cells (CD34+ cells; CD34++ CD71/glycophorin-Alo-int CD45int). Cells were collected in sterile eppendorf tubes in 1 mL of serum-free medium (see below for details).
Cell culture of sorted subfractions.Initially, FACS-sorted cells from maternal PB and control samples were placed on ice for 15 minutes to 2 hours before preparation for fixation and FISH. However, the overall morphology and quality of the preparations, particularly that of the NEC fraction, was found to be compromised by this procedure. Therefore, subsequent sorted samples were cultured in serum-free media before analysis by FISH. For this purpose the purified NECs and CD34+ cells from control and maternal PB samples were centrifuged, resuspended in 100 μL of serum-free media, and split in half for overnight and 5-day cultures. Serum-free medium (Iscove's modified Dulbecco's medium with bovine serum albumin, insulin, transferrin, 2-mercaptoethanol, low-density lipoprotein, and pen-strep)23 was supplemented with combinations of the following hematopoietic cytokines: mast cell growth factor or steel factor (MGF; 50 ng/mL), interleukin-6 (IL-6; 10 ng/mL), granulocyte-macrophage colony-stimulating factor (GM-CSF )/IL-3 fusion protein24 (GM-CSF/IL-3 fusion protein; 20 ng/mL), M-CSF (10 ng/mL), erythropoietin (Epo; 3 U/mL), and Flt-3 ligand (50 ng/mL). MGF, GM-CSF/IL-3 fusion protein, Flt-3 ligand, and IL-6 were kindly provided by Dr D.E. Williams (Immunex, Seattle, WA). M-CSF was kindly provided by Genetics Institute (Cambridge, MA). Epo was provided by colleagues in our laboratory. Cells were cultured in 96-well tissue culture plates (Nunc, Kamstrup, Denmark) for 18 hours and 5 days at 37°C, 5% CO2 in air, in a fully humidified incubator and at a concentration no greater than 50,000 cells/well. In addition, the sorted fractions from women pregnant with trisomic fetuses and 5 of the 10 fresh maternal PB samples were cultured for a total of 21 days and aliquots removed on days 5, 10, and 21 for FISH analysis.
Preparation of slides for FISH.Nucleated erythroid and CD34+ cell fractions from maternal PB and controls were obtained after overnight and 5-day cultures and prepared for FISH. For this purpose, cells were treated with hypotonic solution and placed on poly-l-lysine coated slides (6-mm wells; Cel-Line Assoc, Newfield, NJ). Individual slides (8 slots per slide) contained maternal PB (NEC and CD34+ subfractions) and control FL and PB cells. The cells were fixed to the slides with successive treatments of 20%, 50%, and 100% Carnoy's fixative (3:1 methanol:glacial acetic acid) and air-dried overnight. The slides were stored frozen at −20°C in air-depleted nitrogen-enriched sealed cryopreservation bags until further use. After thawing, the slides were treated with 2× sodium citrate sodium chloride (SSC) for 30 minutes at room temperature and dehydrated in ice-cold 70%, 80%, 90%, and 100% ethanol (2 minutes each) and air-dried. DNA on the slides was denatured in 70% formamide (Life Technologies, Gaithersburg, MD) in 2× SSC for 5 minutes at 70°C, dehydrated as noted above, and dried at 37°C before hybridization.
DNA probes for FISH.Detection of target sequences on human chromosomes X and Y was achieved using probes specific for the alphoid repeat sequence located at the centromere of chromosome X (Xq11.1)25 and the heterochromatic region on the long arm of the Y chromosome.26 The probes were directly labeled by nick translation with either ChromaTide BODIPY-texas red (TR)-X-14-dUTP or BODIPY-fluorescein (FL)-X-14-dUTP (Molecular Probes, Eugene, OR). A Locus Specific Identifier chromosome 21 probe directly labeled with spectrum orange (Vysis, Downers Grove, IL) was used according to the manufacturer's instructions.
FISH.Hybridization was performed according to a protocol based on that of Pinkel et al.27 Ten nanograms of X-TR and Y-FL-X DNA was resuspended in 5 μL of hybridization solution (55% formamide, 10% dextran sulfate, 1× SSC and salmon sperm DNA). The probe was denatured for 5 minutes at 70°C and applied to the slides. After overnight hybridization at 37°C, slides were washed in 60% formamide, 2× SSC at 44°C and in 2× SSC (pH 7.0) at 37°C before counterstaining with 4,6-diamidino-2-phenylindole dihydrochloride (DAPI, 250 ng/mL) and mounting in antifade solution (2.3% (wt/vol) 1,4-diazobicyclo-(2,2,2)-octane (DABCO; Sigma Chemical Co). The interphase and metaphase preparations were visualized with an Axioplan II fluorescent microscope (Zeiss, Thornwood, NY) equipped with two filter sets: a triple band pass filter for detection of DAPI, TR, and FITC and a filter block containing two single pass filters for PI and FITC (Omega, Brattleboro, VT).
Scoring of the hybridization signals was done without previous knowledge of the fetal karyotype or gravida status of the donor. A positive male signal was defined as an interphase nuclei or metaphase spread containing one X and one Y signal. One hundred percent of the intact fixed cells from maternal PB were counted and scored. Previously fixed metaphase preparations from normal male and female donors were included on each slide to establish the sensitivity of each assay. The hybridization efficiency was optimized using FL and adult lymphocyte interphase preparations. The specificity and sensitivity of both X and Y probes was tested on serial dilutions of male and female cells and on replicate samples of pure female and male PB interphase nuclei.
Histochemical analysis of flow-sorted NEC and CD34+ subfractions.Cytocentrifuge preparations stained with May-Grünwald-Giemsa of up to 50,000 sorted cells were used for morphologic analysis of the cells sorted in the NEC and CD34+ fractions. At least 100 cells were analyzed to give a differential.
Statistical analysis.The probability of significant differences between two unpaired groups (ie, fetal karyotype of 46,XX v 46,XY) was determined by the use of the Mann-Whitney U nonparametric test. The Kruskal Wallis ranking test was used for evaluating significant differences between samples from three or more groups (ie, fetal karyotype of 46,XX v 46,XY v nongravida sample). Contingency tables and chi-square analysis were used to determine which cell population was a better predictor of fetal sex and the positive and negative predictive values. The efficacy of using the isolation of fetal cells from the maternal PB for prenatal diagnosis was evaluated in terms of the positive and negative predictive values. Positive predictive value refers to the probability that the fetus is male given that the FACS/FISH method predicts that the fetus is male. Negative predictive value refers to the probability that the fetus is female given that the FACS/FISH method predicts that the fetus is not male. For all statistical comparisons a P value less than .05 was considered statistically significant.
RESULTS
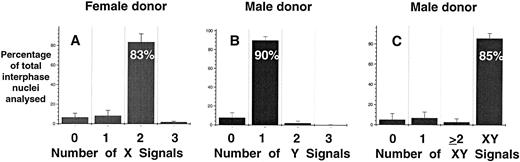
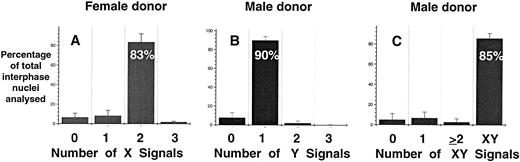
Specificity and sensitivity of FISH analysis.A necessary component of rare-event enumeration is the application of a suitable detection system that is both specific and sensitive. The hybridization efficiency and sensitivity of FISH for the detection of male fetal cells in maternal PB was examined in two independent experiments. Directly labeled probes specific for respective X and Y chromosome were used. In serial dilution experiments, male mononuclear PB cells were artificially mixed with nongravida female mononuclear PB cells (Table 1). In two experiments it was found that 1% male cells could be consistently detected in mixtures of male and female cells. However, it was not possible to consistently detect greater than 91% of the positive events even with 100% pure male or female populations owing to the presence of interphase nuclei with an abnormal number of signals (ie, greater or less than one X and one Y signal or two X signals). The overall specificity of the individual X and Y probes and the two probes combined was 83.5%, 90%, and 85%, respectively, when 100% pure female and male mononuclear cells were analyzed (Fig 1). When both probes were combined and hybridized to only female or male mononuclear cells the false-positive rates (ie, detecting XY or XX signals in female and male preparations, respectively) of the assay were 0.24% and 0.27% (data not shown).
Hybridization efficiency of DNA probes for the detection of X and Y chromosomes in pure populations of female (A) and male (B and C) mononuclear cells. Directly labeled probes specific for X (centromere) and Y repeat sequences were prepared and used as described in Materials and Methods. The results are represented as mean ± SE of eight separate experiments.
Hybridization efficiency of DNA probes for the detection of X and Y chromosomes in pure populations of female (A) and male (B and C) mononuclear cells. Directly labeled probes specific for X (centromere) and Y repeat sequences were prepared and used as described in Materials and Methods. The results are represented as mean ± SE of eight separate experiments.
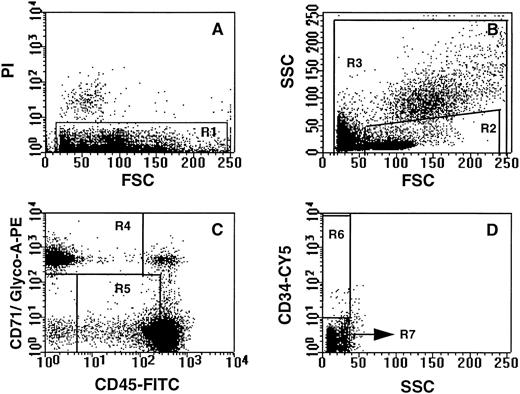
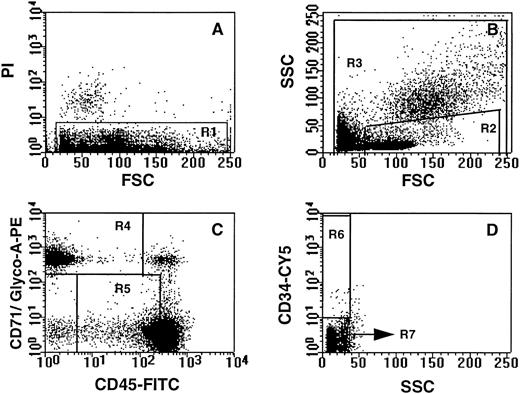
Selection of NECs and CD34+ hematopoietic progenitor cells by flow cytometry and cell sorting.The gating criteria for the isolation of NECs (CD71/glycophorin-A+ CD45lo-int CD34lo) and CD34+ hematopoietic progenitors (CD34++ CD71/glycophorin-Alo-int CD45int) in maternal PB is shown in Fig 2. The gates were selected to recover the highest possible yield of NECs, although a significant population of erythrocytes (glycophorin-A+ CD71lo) was also included in this sort window. However, erythrocytes were lysed during subsequent processing and did not pose a problem in the FISH analysis. Experiments designed to establish a reproducible sorting strategy confirmed that cells sorted within the selected gates can be morphologically identified as nucleated erythroid and blast cells (results not shown).
Sort strategy for the isolation of nucleated erythroid and CD34+ progenitor cell subpopulations. All dot plots are derived from low-density PB cells. Gates for the nucleated erythroid cell population were set to exclude dead (PI+) cells (A; gate R1), events appearing in the last channels of forward light and side scatter (B; gate R3) and CD34+ cells (D; gate R7). A nucleated erythroid cell (NEC) gate was set to include 99% of the CD71/glycophorin-A+ events and cells expressing low to intermediate levels of CD45 (C; gate R4). CD34+ hematopoietic progenitor cells were selected on the basis of viability (A; gate R1), low forward and orthogonal light scatter (B; gate R2), low to intermediate levels of CD71 and glycophorin-A, intermediate levels of CD45 (C; gate R5), and high levels of CD34 (D; gate R6).
Sort strategy for the isolation of nucleated erythroid and CD34+ progenitor cell subpopulations. All dot plots are derived from low-density PB cells. Gates for the nucleated erythroid cell population were set to exclude dead (PI+) cells (A; gate R1), events appearing in the last channels of forward light and side scatter (B; gate R3) and CD34+ cells (D; gate R7). A nucleated erythroid cell (NEC) gate was set to include 99% of the CD71/glycophorin-A+ events and cells expressing low to intermediate levels of CD45 (C; gate R4). CD34+ hematopoietic progenitor cells were selected on the basis of viability (A; gate R1), low forward and orthogonal light scatter (B; gate R2), low to intermediate levels of CD71 and glycophorin-A, intermediate levels of CD45 (C; gate R5), and high levels of CD34 (D; gate R6).
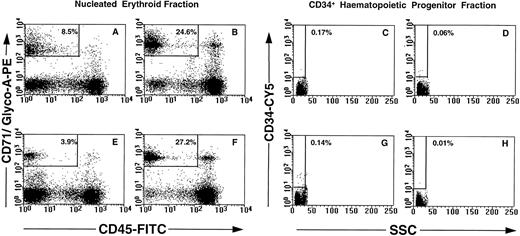
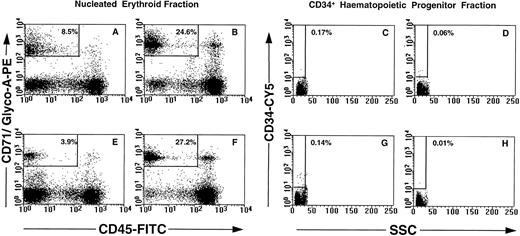
Phenotypic and morphologic analysis of sorted cells.FACS analysis of CD71, glycophorin-A, CD45, and CD34 expression on maternal and nongravida PB mononuclear cells is shown in Fig 3. No significant differences in the overall resolution of the mononuclear cells into distinct subsets was noted between the maternal PB (Fig 3A through D) and the control nongravida PB samples (Fig 3 E through H). The most notable feature is the decrease in the erythrocyte population observed between frozen (Fig 3A and E) and nonfrozen (Fig 3B and F ) PB samples. This resulted in a decrease in the total number of cells falling within the parameters of the NEC gate in frozen PB samples as compared with the nonfrozen specimens (for example, 8.5% v 24.6%, respectively; Fig 3A and B). There was no significant difference in percent CD34+ cells between maternal (Fig 3C and D) and nongravida (Fig 3G and H). Cryopreservation of the mononuclear cells resulted in a slight enrichment in CD34+ cells in both groups (for example, 0.17% v 0.06%, Fig 3C and D).
Multiparameter analysis of previously frozen (A, C, E, and G) and nonfrozen (B, D, F, and H) low-density cells labeled with combinations of MoAbs. Maternal PB (A through D) and nongravida PB (E through H) were used to target two candidate populations of fetal cells: nucleated erythroid cells (NEC; CD71+ glycophorin A+ CD45lo-int CD34− PI−; A, B, E, and F ) and fetal CD34+ progenitor cells (CD34+ CD71− glycophorin A− CD45int PI−; C, D, G, and H).
Multiparameter analysis of previously frozen (A, C, E, and G) and nonfrozen (B, D, F, and H) low-density cells labeled with combinations of MoAbs. Maternal PB (A through D) and nongravida PB (E through H) were used to target two candidate populations of fetal cells: nucleated erythroid cells (NEC; CD71+ glycophorin A+ CD45lo-int CD34− PI−; A, B, E, and F ) and fetal CD34+ progenitor cells (CD34+ CD71− glycophorin A− CD45int PI−; C, D, G, and H).
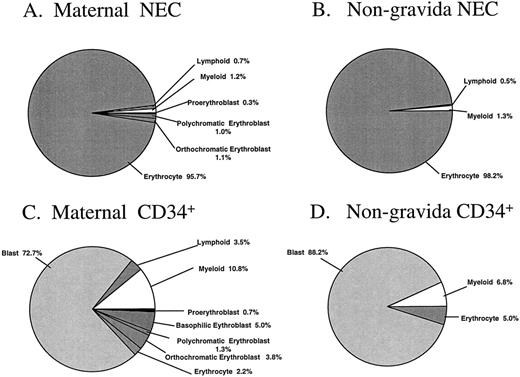
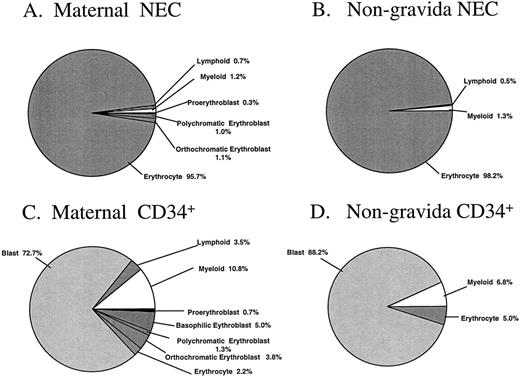
The percentages of morphologically recognizable cells in the sorted cell fractions are summarized in Fig 4A through D. Subtle differences between sorted PB cells from pregnant and nonpregnant women were observed. Nucleated erythroblasts were identified unequivocally in two of the three maternal NEC fractions analyzed, with this cell type representing 2.4% of the total sorted sample (Fig 4A). No attempt was made to distinguish the possible fetal or adult origin of the erythroid cells in these particular samples. There were no erythroblasts identified in the NEC sorted fraction from the nongravida PB samples (n = 4, Fig 4B). The majority of the cells from maternal and nongravida sorted NEC fractions were erythrocytes (95.7 and 98.2%, respectively). The majority of the cells in the CD34+ fractions were blast cells, representing 72.7% and 88.2% of the total cells that were sorted from maternal and nongravida PB, respectively (Fig 4C and D).
Morphologic analysis of the NEC and CD34+ sorted subfractions from previously frozen maternal and nongravida PB samples. Fractions were sorted by FACS and cytocentrifuged at 500 rpm for 5 minutes onto slides and differentially stained with May-Grünwald-Giemsa. No less than 100 cells were analyzed by light microscopy to give a differential. The mean values from three and four independent experiments from maternal and nongravida PB, respectively, are presented.
Morphologic analysis of the NEC and CD34+ sorted subfractions from previously frozen maternal and nongravida PB samples. Fractions were sorted by FACS and cytocentrifuged at 500 rpm for 5 minutes onto slides and differentially stained with May-Grünwald-Giemsa. No less than 100 cells were analyzed by light microscopy to give a differential. The mean values from three and four independent experiments from maternal and nongravida PB, respectively, are presented.
FISH analysis of cells sorted from frozen maternal and nongravida PB samples.PB samples from 27 pregnant women carrying normal 46,XY or 46,XX fetuses were studied for the presence or absence of male fetal cells using FISH. Four previously frozen PB samples from nongravida female donors were also included in the blind study as controls. Of the original 200 expectant mothers who donated 20 mL of PB before amniocentesis for cryopreservation and banking, four were identified as carrying trisomic fetuses (47,XY,+18 and 47,XY,+21). These four PB samples were also included in this study.
In Tables 2-7 results of FISH analysis of the cells recovered from the various sort fractions before and after culture from maternal PB samples (10 to 13 weeks' gestation) are grouped according to their corresponding fetal karyotype (fetal karyotype 46,XY n = 20; 46,XX n = 7; 47,XY,+21 n = 3; and 47,XY,+18 n = 1). The absence of a detectable Y chromosome in the sorted cells (as would be expected with a female fetus) was labeled as “not applicable” in the fetal sex prediction column (n/a; column 7, Tables 2-7) because differentiation between the mother's cells and that of her female fetus was in those instances not possible.
The hybridization efficiency for each maternal PB fraction examined was determined by including control 100% male and female interphase nuclei on the same hybridization slide. This efficiency varied to some extent from experiment to experiment (column 6, Tables 2-7). There was also considerable variation in the number of cells that were recovered after cryopreservation, sorting, and subsequent FISH analysis between the different maternal PB samples analyzed.
FISH analysis of cell flow sorted within the NEC window.Table 2 outlines the combined results (18-hour and 5-day cultures) of FISH analysis of the NEC fraction from previously frozen maternal PB (n = 29) and nongravida control samples (n = 4). Between 408 and 52,134 cells were sorted from the frozen PB samples (column 3, Table 2). Among the 29 sorted samples, pregnancies involving normal male fetuses were correctly identified in 6 of 19 (32%) cases. Thus, there were no fetal cells detected in 13 of the 19 informative NEC fractions analyzed. The range of the frequency of fetal cells in the NEC fraction was 0 to 14 (column 5, Table 2). The false-positive rate was 33% (ie, XY cells were detected in 2 of 6 maternal PB NEC fractions analyzed with a fetal karyotype of 46,XX). Both false-positive results were from PB samples from mothers who previously gave birth to male children. In addition, two of the four sorted fractions from nongravida control donors contained one XY interphase nuclei each. Using a Mann-Whitney nonparametric test to determine significance when evaluating male (XY) cells found when the fetal karyotype was truly XY (true positive) compared with when the fetal karyotype was XX and from nongravida samples (false positive), it was found that there was a significant difference between the two groups (P = .05). Two of the three NEC fractions from maternal PB in which the fetal karyotype was 47,XY,+21 (samples numbered 42 through 44) were informative not only for the prediction fetal sex, but also for the detection of trisomy 21.
FISH analysis of cells sorted within the CD34+ window.The results of FISH analysis of CD34+ cells sorted from 28 maternal PB samples and cultured for 18 hours and 5 days are summarized in Tables 3 and 4, respectively. Between 372 and 23,800 CD34+ cells were sorted, split into two fractions, and cultured for 18 hours (overnight) and 5 days (Column 3, Table 3 and 4). In the maternal PB fractions examined in which the corresponding fetal karyotype was 46,XY, the gender prediction accuracy was 42% (8 of 19) and 44% (8 of 18) in the overnight and 5-day cultures, respectively. The range of the detectable XY CD34+ cells was 0 to 35 cells per 20 mL (2 × 107 cells) of maternal PB originally frozen. In 3 of the 5-day cultured maternal PB CD34+ sorted samples (XY fetal karyotype) analyzed, at least 1 male metaphase was detected. In 11 of 19 (58%) and 10 of 18 (55%) fractions analyzed from the overnight and 5-day cultures, respectively, there were no XY cells detected, although the fetal karyotype was XY. The false-positive rate for the CD34+ fraction cultured overnight was 1 of 5 in maternal PB samples (fetal karyotype 46,XX) and 1 of 4 in the nongravida samples. The false-positive rate for the CD34+ fractions cultured for 5 days from the group of 6 women carrying a female fetus and four non-gravida donors was 1 of 6 and 1 of 4, respectively. In the CD34+ fraction cultured overnight there were male cells detected in all of the maternal PB examined in which the fetus was trisomic. The range of the frequency of these cells was from 1 to 9 detected cells per 20 mL of blood. Three of the four 5-day cultures of CD34+ cells from mothers expecting a trisomic fetus yielded between 3 and 26 male cells, whereas there were no male cells detected in the fourth sample (Table 4, sample no. 45). Interestingly, when CD34+ cells from these mothers were kept in culture for a total of 3 weeks and then subsequently analyzed by FISH, 5 male cells were detected in the maternal PB sample no. 45 (data not shown).
When the results from all three cell fractions and culture periods were pooled, pregnancies involving normal male fetuses were correctly identified by FISH in 65% (13 of 20) of the cases. The overall frequency of male fetal cells varied from 0 to 35 per 2 × 107 (20 mL) of frozen PB cells. The false-positive rate was 3 of 7 and 2 of 4 for PB from pregnancies involving a female fetus and nongravida donors, respectively. Chi-squared analysis of the XY cells detected in the sorted cell fractions showed that the CD34+ fraction was a better predictor of fetal sex (P = .20). The combined positive and negative predictive values were 84% and 26%, respectively. The gender of the trisomic fetus was accurately predicted in all four of the associated maternal PB analyzed. In all 3 of the trisomy 21–associated maternal PB analyzed, the trisomic fetal karyotype was confirmed using the chromosome 21–specific probe. The number of fetal trisomic cells ranged from 0 to 26 cells recovered from 20 mL of PB.
FISH analysis of nonfrozen maternal PB samples.It was of interest to determine if the cryopreservation of maternal PB samples had any detrimental effect on the recovery of fetal cells. This was of particular concern in the NEC fraction, which contains fragile lineage-committed and terminally differentiated erythroid cells. Ten freshly Ficolled, nonfrozen maternal PB samples were enriched for NECs and CD34+ hematopoietic progenitor cells by flow cytometry and cell sorting as before and analyzed for the presence of XY cells by FISH (Tables 5-7). The recovery of erythroid cells from the nonfrozen samples was higher than was observed in the frozen maternal PB samples. The number of cells recovered from the NEC fraction varied from 1.7 × 105 to 1.2 × 106 (Table 5). Fetal cells were ultimately detected in the NEC fraction in 3 of the 7 maternal PB analyzed in which the fetal karyotype was XY. The range of the frequency of these cells was from 1 to 68. One of the NEC fractions from a mother carrying a female fetus contained 2 XY cells. The gender prediction accuracy was higher in the CD34+ fractions, although the false-positive rate also increased. Between 0 and 93 XY cells were detected in the two CD34+ cultured fractions (Tables 6 and 7). Overall, when the results from the NEC and the CD34+ fractions are combined, male cells were detected in all of the fresh maternal PB samples examined. When analyzed this way there were two false-positive results from three maternal PB (fetal karyotype 46,XX) samples analyzed.
A summary of the frequencies of fetal cells isolated from previously frozen and nonfrozen maternal PB and the positive and negative predictive value of these analyses as predictors of fetal karyotype is displayed in Table 8.
DISCUSSION
The isolation of fetal hematopoietic cells from the maternal circulation has the potential to allow early prenatal diagnosis without having to invade the gravid uterus. This represents an enormous challenge given the rarity of these cells. Indeed, considerable progress has been made in the qualitative assessment of fetal cell populations found in maternal PB. However, more information is needed regarding the enumeration and characterization of fetal cell target populations. This report describes our efforts to quantitatively assess and compare simultaneously two phenotypically well-defined candidate fetal cell populations from previously frozen and nonfrozen maternal PB. CD34+ hematopoietic progenitor and nucleated erythroid candidate fetal cell subpopulations were enriched by FACS sorting on the basis of forward and orthogonal light scatter properties and the expression of CD71, CD45, CD34, and glycophorin-A. The sorted subpopulations were analyzed morphologically and the majority of the cells were immature blast cells and cells at different stages of erythroid differentiation. The fetal origin of the sorted cells was confirmed using FISH and probes specific for chromosomes X, Y, and 21.
Nucleated erythroblasts represent an attractive fetal cell population to target because these cells are abundant in the fetus and rare in the periphery of normal healthy adults. In addition, these cells have a limited life span: 8 days from the proerythroblast to reticulocyte stage. Other groups have documented the isolation of fetal nucleated “erythrocytes” using FACS4,14,15,21,22 or magnetic cell sorting16,28 by positive or negative selection. However, a consensus on the expected frequency and the best method to retrieve these fetal cells in normal pregnancies has not been reached. In the present study, male fetal cells with a CD71/glycophorin-A+ CD45lo-int CD34lo phenotype were detected in only 32% (6 of 19) of the fractions analyzed and at a frequency of 0 to 14 cells per 2 × 107 (20 mL) cryopreserved maternal PB cells. The observed frequency of erythroblasts of fetal origin in our study is compatible with other reports.4,28,29 Morphologic examination of the NEC fraction sorted from frozen maternal PB showed that the majority of NECs were terminally differentiated (polychromatic and orthochromatic) erythroblasts. These cells are fragile and presumably more susceptible to fracture caused by freezing. Indeed, the yield of fetal NECs was greater in the NEC fractions from nonfrozen PB samples (frequency range = 0 to 68 per 20 mL of maternal PB); however, NECs were detected in only 43% of the maternal PB examined (fetal karyotype 46,XY). Therefore, the vast majority of cells that were morphologically identified as erythroblasts (2.4%; Fig 4A) must have been of maternal origin. These results are in keeping with those reported by others.13,30 During pregnancy, stimulation of maternal erythropoiesis parallels the observed increases in blood volume and oxygen demand.31 Furthermore, it has been shown that blood from pregnant females contain large numbers of erythroid progenitors (BFU-E).13 The presence of maternally derived erythroblasts in normal maternal PB documented in this study may explain reports of abundant “fetal nucleated erythrocytes” in previous investigations in which the fetal origin of the erythroblasts was not determined.16 17
Given the limited life span of erythroblasts and the many reports indicating detection of fetal cells from day 33 to 16 weeks of gestation, it is assumed that the first trimester leakage of erythroid cells from the fetal/placental barrier is a continuous process involving minute quantities of fetal cells. Therefore, it is unlikely that the “window of opportunity” was missed in this study of women at 10 to 13 weeks' gestation. In addition, it is unlikely that fetal cells were lost because of cell clumping.29 RBC agglutination attributed to the glycophorin-A MoAb was not a problem in our experiments, most likely because of the low frequency of RBCs in (previously frozen) mononuclear cell fractions obtained by Ficoll density centrifugation. Although the passage of fetal NECs from the fetal/placental unit to the mother may represent a normal physiologic event in all pregnancies and may play a role in inducing tolerance to the fetal allograft as has been suggested by others,20 the results presented here illustrate that the frequency of such cells in relatively small volumes of maternal PB (20 mL) poses a serious technical challenge that was only partly met by the technologies applied in our study.
Previous studies have shown that fetal cells can be detected within the CD34+ compartment of PB from expectant mothers.19 20 These observations were confirmed and extended in this study. Fetal hematopoietic precursors with a CD34++ CD71/glycophorin-Alo-int CD45int phenotype were detected in 42% (8 of 19) of the PB fractions from mothers carrying a normal male fetus. The range of the frequency of these cells was similar to that observed the NEC fractions (0 to 7 fetal cells). However, a slight expansion in the frequency of these primitive cells was observed in the CD34+ fraction that was cultured for 5 days (0 to 35 fetal cells). This expansion was also noted in CD34+ fractions isolated from nonfrozen maternal PB and cultured for 5 days. The recovery of total CD34+ cells from nonfrozen maternal PB was higher (mean, 7,480 cells) than from previously frozen samples (mean, 5,148 cells) and similarly more male fetal cells were identified in some of the nonfrozen fractions (0 to 93 male fetal cells detected per 20 mL [2 × 107 cells] of maternal PB). Interestingly, in 3 of the 18 cultured (previously frozen) maternal PB CD34+ sorted samples (fetal karyotype 46,XY), at least 1 male fetal metaphase was detected. This observation indicates that (some of ) these fetal blood cell precursors are mitotically active, thus giving renewed hope to the possibility of clonally expanding isolated fetal cells. The clonal expansion of fetal cells is highly desirable given the rarity of these cells and the advantages associated with replicate genetic analysis.
The cytokine combination used in this study has been previously shown to be capable of supporting and sustaining FL-derived hematopoietic cells.32 33 Other, perhaps yet unidentified (combinations of ) factors may induce a more vigorous proliferative response, promoting the in vitro expansion of fetal cells isolated from maternal blood. This possibility warrants further study.
Although the FISH assay had a high sensitivity and specificity and precautions were taken to avoid contamination of the preparations with cells from other sources, false-positive and -negative results were detected in all the fractions analyzed. These spurious results could be the result of nonspecific binding of the Y probe to nontarget sequences, the inability to detect one of the X signals (ie, the resolution of signals on different focal planes), or the contamination of male cells from different sources. The possibility that the false-positive results from the CD34+ hematopoietic progenitor fraction are residual fetal cells from past pregnancies was not examined in this study but certainly could explain some of our observations. The vast majority of women who volunteered for this study were woman of advanced maternal age and multigravida status. The detection of true positives may be increased and false positives decreased in future studies by using synthetic peptide nucleic acid oligonucleotide probes for FISH, which we have recently shown to be more specific and efficient than conventional DNA probes.34 In terms of false-negative results, it should be noted that the estimated frequency of fetal cells in our study could have underestimated the actual frequency of fetal cells in maternal blood. Significant numbers of cells may have been lost in both the cell selection process and the subsequent in situ hybridization procedure (see, eg, number of cells sorted v number of cells analyzed by FISH; columns 3 v 4 in Tables 2-7).
Based on previous reports,4,10 11 it was anticipated that large numbers of fetal cells would be detected in pregnancies involving a triploid fetus. Although fetal cells were ultimately observed in 2 of the 4 NEC and 4 of the 4 CD34+ fractions analyzed, the number of enriched fetal cells was not significantly higher than those observed in normal pregnancies. The possibility that the detected CD34+ fetal cells were cells from previous pregnancies can be excluded in these instances because this was the first pregnancy involving a trisomic fetus for all of the women studied. Additional studies involving aneuploidy pregnancies are needed to determine if the observed frequencies are associated with a more permissive placenta and more consistent deportation of fetal cells resulting in a higher detection rate compared with normal pregnancies.
When the results from all three subfractions and culture times are pooled, male fetal cells were detected in 65% (13 of 20) of PB from women pregnant with normal male fetuses. The frequency of male fetal cells varied from 0 to 35 per 2 × 107 (equivalent to 20 mL) of frozen PB cells. The gender and the trisomy of the fetus was accurately predicted from the isolated fetal cells in all three of the associated maternal PB analyzed. Overall, fetal cells were identified in all of the nonfrozen PB fractions analyzed in which the fetal karyotype was 46,XY. Less than 100 fetal cells were detected in 2 × 107 (20 mL) nonfrozen maternal PB mononuclear cells. These results are encouraging and provide some useful information regarding the frequency and the characterization of potential fetal cell target populations. However, at the present time, the isolation of fetal cells from the maternal PB may only provide an important adjunct to the current noninvasive “triple marker” screening. More research into methods for the enrichment and identification of these rare cells needs to be completed before fetal cells isolated from the maternal periphery will become a noninvasive diagnostic reality.
ACKNOWLEDGMENT
Gayle Thornbury is thanked for the FACS analysis and cell sorting. Excellent technical assistance was provided by Coleen McAloney and Cam Smith. The assistance from Gloria Shaw and Donna Hogge with FISH is gratefully acknowledged. MGF, IL-6, Flt-3 ligand, and GM-CSF/IL-3 fusion protein were generously provided by Immunex. M-CSF was a gift from Genetics Institute. Ethylin Wang Jabs provided the Xc DNA clone. Sara Abraham, Heather Kirk, and Terry Thomas are thanked for critically reviewing the manuscript. We also thank the volunteers who donated PB for this study.
Supported by a grant from the Medical Research Council of Canada and was presented in part at the American Society of Hematology conference, Seattle, WA, December 1-5, 1995.
Address reprint requests to Peter M. Lansdorp, MD, PhD, Terry Fox Laboratory, 601 W 10th Ave, Vancouver, BC Canada V5Z 1L3.