Abstract
The RET proto-oncogene product is a receptor tyrosine kinase representing the signal-transducing molecule of a multisubunit surface receptor complex for the glial cell line-derived neurotrophic factor (GDNF ), in which a novel glycosyl-phosphatidylinositol (PI)-linked protein (termed GDNFR-α) acts as the ligand-binding component. We have analyzed expression of RET and GDNFR-α in purified normal hematolymphopoietic cells, leukemia/lymphoma cell lines, and 154 primary samples from patients with hematopoietic malignancies encompassing different lineages and differentiation stages. Relatively low amounts of RET mRNA were found in early CD34+ hematopoietic progenitors, but RET transcripts appeared to increase after myelomonocytic maturation. No expression of RET was found in peripheral blood and tissue B and T lymphocytes. Analysis of human myelomonocytic cell lines was overall consistent with results obtained on purified normal cells. Accordingly, RET expression was mainly confined to acute myeloid leukemias (AMLs) displaying either monocytic (French-American-British M4 and M5) or intermediate-mature myeloid (M2 and M3) phenotypes, being less frequently detected in early myeloid (M0 and M1) AMLs. In contrast, RET mRNA was sporadically detected in B-cell tumors, whereas, among T-cell malignancies, RET transcripts were mainly detected in cells of postthymic and mature T-cell phenotype. RET broad detection in primary tumors was not paralleled by the mutual expression of GDNFR-α, which was detected only in 2 isolated primary samples and in 3 leukemia/lymphoma cell lines. However, GDNFR-α transcripts, in the absence of RET mRNA, were found in normal bone marrow stromal cells (BMSC), in BM fibroblasts, and in two osteoblast cell lines previously described to support normal hematopoiesis. In the presence of GDNF-receptors derived from BMSC by PI-specific phospholipase C cleavage, GDNF efficiently bound RET-expressing AML blasts and was functionally active by reducing their clonogenic growth and triggering the monocytic maturation of leukemic cells.
HEMATOPOIESIS IS A tightly regulated process in which a small population of self-renewing primitive progenitors generates an offspring of increasingly differentiated end cells with specific functional activities.1 This process is controlled by a number of growth factors and cytokines,1 with some of them exerting their specific functions through the binding to high-affinity receptor tyrosine kinases (RTKs), which are differentially expressed on the various hematopoietic cell subsets.2,3 RTKs have been shown to act as important regulators in the processes of growth and differentiation of hematopoietic progenitors. For example, the type III RTK subfamily includes the product of proto-oncogene FMS, which was identified as the receptor for macrophage colony-stimulating factor,4 a critical cytokine for cells of the monocyte-macrophage1,4 and osteoclast lineages.5 Similarly, products of KIT and FLT3/FLK2 genes, respectively representing transmembrane receptors for stem cell factor6 and FLT3-ligand,7,8 are other type III RTKs playing a pivotal role in the early steps of hematopoiesis and contributing to the functional regulation of specific cell types, such as CD34+ progenitors, mast cells, megakaryocytes, and osteoclasts.6,8,9 More recently, a novel RTK, named TNK1, exerting a regulatory role in early hematopoiesis, has been cloned from human umbilical cord blood CD34+ stem cells.10 In addition, the product of TRK proto-oncogene, encoding a high-affinity nerve growth factor receptor, although first described as a nonhematopoietic RTK, has been later shown to be expressed and functionally active in normal and malignant myelomonocytic cells.11
The RET proto-oncogene encodes for a member of the RTK superfamily2 whose structure consists of cadherin-like and cysteine-rich repeats in the extracellular region, a hydrophobic transmembrane domain, and a split intracellular tyrosine kinase region.12-14 Recently, evidence has been provided that RET may act as a signaling component of a multisubunit receptor complex for the glial cell line-derived neurotrophic factor (GDNF ),15,16 a potent neutrophic factor acting on central and peripheral neurons.17,18 Independent studies have shown that GDNF binds to a novel glycosylphosphatidylinositol (GPI)-linked protein (termed GDNFR-α) by promoting the formation at the cell surface of a physical complex involving GDNF, GDNFR-α, and RET.19 20 As a result of such interactions, RET-mediated intracellular signaling is activated.19 20
Although previous studies have indicated an important role for the RET product in the physiologic development and differentiation of neural crest derivatives, enteric nervous system, and components of the excretory system,21-25 as well as its involvment in some forms of human neoplasia,26,27 the role of this RTK in human hematopoiesis has not been addressed so far in detail. On the other hand, RET transcripts and/or protein have been detected in hematopoietic fetal liver22 and in two myeloid leukemic cell lines of human origin, ie, HL-60 and THP-1,28-30 thus suggesting that some cells of hematopoietic origin could express the RET-encoded RTK.
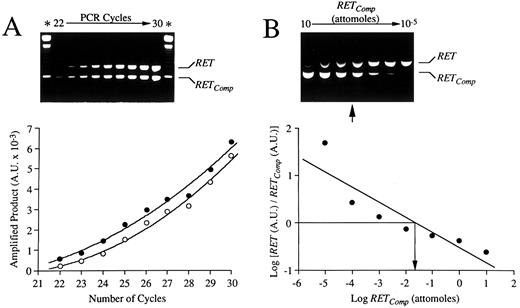
Validation of competitive RT-PCR strategy by use of a nonhomologous DNA fragment (RETComp ) engineered to contain specific RET primer templates. (A) Kinetics of amplification of RET cDNA and RETComp fragments. Fixed amounts (0.1 attomoles) of RET cDNA and RETComp fragments were amplified in a single reaction tube with specific RET primers. After 22 amplification cycles and after each of 8 additional cycles, a small aliquot of the reaction mixture was removed and the products were resolved on agarose gel (upper panel). The relative intensities of the bands corresponding to RET cDNA (790 bp) and RETComp (597 bp) -amplified products were quantified by computer imaging. The amount of specific amplified products for RET cDNA (○) and RETComp (•), expressed in AU, was plotted as a function of the number of cycles (lower panel). (B) Determination of relative levels of RET mRNA in THP-1 cells by competitive RT-PCR. Ten-fold serial dilutions (10 to 1 × 10−5 attomoles) of RETComp were amplified with RET primers together with constant aliquots of cDNA from the THP-1 cell line. After 35 cycles, amplified products were resolved on agarose gel (upper panel). Relative intensities of the bands were densitometrically determined and the logarithm of their ratios was plotted as a function of the logarithm of the amount of RETComp added (lower panel). The equivalence point (arrow) was inferred between 10−1 and 10−2 attomoles.
Validation of competitive RT-PCR strategy by use of a nonhomologous DNA fragment (RETComp ) engineered to contain specific RET primer templates. (A) Kinetics of amplification of RET cDNA and RETComp fragments. Fixed amounts (0.1 attomoles) of RET cDNA and RETComp fragments were amplified in a single reaction tube with specific RET primers. After 22 amplification cycles and after each of 8 additional cycles, a small aliquot of the reaction mixture was removed and the products were resolved on agarose gel (upper panel). The relative intensities of the bands corresponding to RET cDNA (790 bp) and RETComp (597 bp) -amplified products were quantified by computer imaging. The amount of specific amplified products for RET cDNA (○) and RETComp (•), expressed in AU, was plotted as a function of the number of cycles (lower panel). (B) Determination of relative levels of RET mRNA in THP-1 cells by competitive RT-PCR. Ten-fold serial dilutions (10 to 1 × 10−5 attomoles) of RETComp were amplified with RET primers together with constant aliquots of cDNA from the THP-1 cell line. After 35 cycles, amplified products were resolved on agarose gel (upper panel). Relative intensities of the bands were densitometrically determined and the logarithm of their ratios was plotted as a function of the logarithm of the amount of RETComp added (lower panel). The equivalence point (arrow) was inferred between 10−1 and 10−2 attomoles.
To better understand whether RET receptor might be involved in the regulation of human hematopoiesis, we have analyzed its levels of expression in purified normal and malignant cells of myeloid and lymphoid lineages mirroring various stages of hematolymphopoietic differentiation, leukemia/lymphoma cell lines, and stromal cells of the bone marrow microenvironment. In addition, the presence of GDNFR-α in the same cell types and the functional effects of GDNF on human leukemic cells were analyzed.
MATERIALS AND METHODS
Cell samples.The study included cellular samples obtained from peripheral blood (PB) or bone marrow (BM) of 154 patients with acute myeloid leukemias (AML; n = 53); chronic myeloproliferative disorders in myeloid blast crisis (MBC; n = 5); B- and T-cell lymphoproliferations (n = 96), including B- and T-lineage acute lymphoblastic leukemias (ALL); chronic lymphocytic leukemia (CLL); prolymphocytic leukemia (PLL); hairy cell leukemia (HCL); high- and low-grade non-Hodgkin's lymphomas (NHL) in overt leukemic phase; multiple myeloma (MM); and adult T-cell leukemia/lymphoma (ATLL). Diagnoses were based on cell morphology, immunophenotyping, enzyme cytochemistry, and clinical parameters. Acute leukemias were classified according to the revised French-American-British (FAB) criteria.31 NHL was diagnosed by histopathologic examination of lymph node tissues and immunohistochemistry and classified according to the International Working Formulation.32
Cell isolation and purification.Anticoagulated PB and BM aspirates were collected from leukemia/lymphoma patients after informed consent was obtained and before therapy. Neoplastic cells were isolated by centrifugation on a Ficoll-Hypaque (Pharmacia, Uppsala, Sweden) gradient and, with the exclusion of T-cell malignancies, further purified by T-cell depletion with anti-CD2 immunomagnetic beads (Dynabeads; Dynal, Oslo, Norway). In the case of MM, tumor cells were further purified by positive indirect immunomagnetic selection with the plasma cell-specific monoclonal antibody (MoAb) BB-4.33 After purification procedures were performed, all of the samples contained more than 95% of neoplastic cells. Purification of normal cells from PB and tissues was performed essentially as described.34,35 Briefly, Ficoll-Hypaque–isolated circulating mononuclear cells were further purified by positive immunomagnetic selection with anti-CD2, anti-CD19, anti-CD4, and anti-CD8 immunomagnetic beads (Dynal) to obtain, respectively, T and B lymphocytes as well as CD4+ and CD8+ T-cell subsets. For in vitro activation studies, purified CD2+/CD3+ T lymphocytes were exposed to 12-O-tetradecanoylphorbol-13-acetate (TPA; Sigma, St Louis, MO; 10 ng/mL) plus ionomycin A (Sigma; 1.0 μg/mL) for 72 hours, as reported.34,35 Plastic-adherent macrophages were recovered after 2 hours of incubation at 37°C of the CD2−/CD19− PB mononuclear cell (PBMC) fraction. In some experiments, adherent macrophages were activated in vitro by exposure to 100 ng/mL of granulocyte-macrophage colony-stimulating factor (GM-CSF ) for 12 hours. More than 96% pure neutrophils and eosinophils were isolated from PB buffy coats by 1.2% dextran sedimentation, followed by a multiple density Percoll gradient centrifugation, as described.36 Tissue T and B lymphocytes were purified by tearing out single cells from freshly excised tonsils and thymectomy samples. After Ficoll-Hypaque separation, CD2+, CD4+/CD8+, and CD19+ cells were further purified by immunomagnetic selection. CD34+ hematopoietic progenitors were isolated from PB of patients undergoing high-dose chemotherapy followed by granulocyte colony-stimulating factor and from umbilical cord blood by using affinity columns (MACS CD34 Cell Isolation Kit; Miltenyi Biotec, Celbio, Milan, Italy).36 In all experiments, purity of the selected cell fractions was estimated by morphology and flow cytometry by appropriate MoAb combinations.34 BM aspirates, which were obtained after informed consent from patients with solid tumors undergoing routine staging procedures, were used as a source of BM-derived primary stromal cells (BMSC) and fibroblasts (BMF ). BMSC were obtained from Ficoll-Hypaque–separated mononuclear BM cells cultured in Iscove's modified Dulbecco's medium (IMDM) supplemented with 12.5% fetal calf serum (FCS), 12.5% horse serum (Hyclone, Logan, UT), and 1.0 × 10−6 mol/L hydrocortisone hemisuccinate (Sigma), as previously described.9 For BMF preparation, hydrocortisone was substituted for with 0.1 ng/mL basic fibroblast growth factor (bFGF; Genzyme Co, Cambridge, MA). After four to five passages, cultures developed in the presence of bFGF were virtually free of contaminating endothelial cells and macrophages, as verified by immunostaining for von Willebrand's factor, nonspecific esterase, and CD14.9
Constitutive expression of RET transcripts (semiquantitative RT-PCR) and GDNFR-α (RT-PCR) in normal lymphohematopoietic cells. Cell fractions were isolated to purity by discontinuous gradient centrifugation and/or immunomagnetic selection. Peripheral T cells were activated with 10 ng/mL TPA plus 1.0 μg/mL of ionomycin A for 72 hours. Adherent macrophages (Adh. macroph.) were activated with 100 ng/mL of GM-CSF for 12 hours. In all cases, 1 μg of total RNA was reverse-transcribed in a 20-μL reaction mix containing hexadeoxyribonucleotides random primers. Four microliters was amplified with primers specific for RET and GDNFR-α. In the case of RET, amplification was performed in the presence of a constant amount (10−3 attomoles) of RETComp fragment. After 35 cycles of amplification, 15 μL of PCR products was resolved on 1.5% agarose gel, blotted, and hybridized with specific RET and GDNFR-α oligoprobes. For semiquantitative evaluation of RET transcripts, ratios of the relative intensities of bands corresponding to RET cDNA (790 bp) and RETComp (597 bp) -amplified products were quantified by computer imaging of gel, expressed in AU, and graphed. An adult human brain substantia nigra cDNA and cDNA derived from the MN-60 cell line were respectively used as positive (+) and negative (−) controls for RET and GDNFR-α expression. cDNAs were always tested with β-actin–specific primers.
Constitutive expression of RET transcripts (semiquantitative RT-PCR) and GDNFR-α (RT-PCR) in normal lymphohematopoietic cells. Cell fractions were isolated to purity by discontinuous gradient centrifugation and/or immunomagnetic selection. Peripheral T cells were activated with 10 ng/mL TPA plus 1.0 μg/mL of ionomycin A for 72 hours. Adherent macrophages (Adh. macroph.) were activated with 100 ng/mL of GM-CSF for 12 hours. In all cases, 1 μg of total RNA was reverse-transcribed in a 20-μL reaction mix containing hexadeoxyribonucleotides random primers. Four microliters was amplified with primers specific for RET and GDNFR-α. In the case of RET, amplification was performed in the presence of a constant amount (10−3 attomoles) of RETComp fragment. After 35 cycles of amplification, 15 μL of PCR products was resolved on 1.5% agarose gel, blotted, and hybridized with specific RET and GDNFR-α oligoprobes. For semiquantitative evaluation of RET transcripts, ratios of the relative intensities of bands corresponding to RET cDNA (790 bp) and RETComp (597 bp) -amplified products were quantified by computer imaging of gel, expressed in AU, and graphed. An adult human brain substantia nigra cDNA and cDNA derived from the MN-60 cell line were respectively used as positive (+) and negative (−) controls for RET and GDNFR-α expression. cDNAs were always tested with β-actin–specific primers.
Cell lines and culture conditions.K562 (early myeloerythroid), HEL (myeloblastic-erythroblastic), KG-1A (early myeloblasts, CD34+), KG-1 (early myeloblasts, CD34−), HL-60 (intermediate myeloid-promyelocytes), U937 (early monoblasts), ML3 (myelomonoblasts), THP-1 (monoblasts), Mo7e (megakaryoblast), NB-4 [leukemic promyelocytes harboring the t(15; 17) translocation], FLG 29.1 (pre-osteoclasts), Molt-4, FRO (common thymocyte phenotype, T cells), Jurkat, CEM, KE37 (postthymic phenotype, T cells), H9, HUT 78, HUT 102, Karpas 299 (mature T-cell phenotype), BV-173 (early B lymphoblasts; lymphoid blast crisis of chronic myelogenous leukemia), Ci-1, Ri-1, SC-1 (B-cell NHL), Nalm-6 (early pre-B cells), MN-60 (SIg+ B-cell ALL), HBL-1, HBL-2, HBL-3 (small noncleaved cell lymphoma from human immunodeficiency virus [HIV]+ patients), JD38, Namalwa (sporadic and endemic Burkitt lymphoma), U266, LP1, IM9 (MM), Saos-2, MG-63 (osteoblast cells), and C433 (derived from the stromal component of a giant cell tumor of bone) cell lines were cultured in RPMI 1640 medium (GIBCO, Paisley, UK) supplemented with 10% of FCS, with the exception of KG-1, HEL, and THP-1, which were maintained in IMDM (GIBCO) plus 20% FCS and Saos-2 and MG-63, cultured in McCoy's medium (GIBCO) supplemented with 10% FCS. Mo7e cells were cultured in IMDM plus 5% FCS supplemented with 10 ng/mL GM-CSF. Sources and phenotypic characterization of all the above cell lines have been reported in detail previously.9,34,35,37 U937 cells were induced to differentiate into adherent mature macrophages by incubation with 1 × 10−7 mol/L TPA or vitamin D3 (250 ng/mL; kindly supplied by Dr J. Hadvary, Hoffmann-La Roche, Basel, Switzerland) plus transforming growth factor-β (TGF-β; 1.0 ng/mL; R&D System Europe, Abington, UK).38
RNA extraction and reverse transcriptase-polymerase chain reaction (RT-PCR).Total RNA (1 μg), extracted by the guanidium thiocyanate method,39 as well as poly A+ RNA from human adult brain substantia nigra (Clontech Laboratories Inc, Palo Alto, CA) were reverse-transcribed by avian myeloblastosis virus reverse transcriptase (Promega, Madison, WI) for 1.5 hours at 42°C in a 20-μL reaction mix containing hexadeoxyribonucleotides random primers (0.4 μg). Four microliters of the same cDNA preparations was amplified in a 50-μL volume of final reaction mix in a Perkin Elmer 9600 thermal cycler (Perkin Elmer Cetus, Emeryville, CA) with 25 pmol of primers specific for RET (sense, 5′-CAG CTG CTT GTA ACA GTG-3′, region 1426-1443; antisense, 5′-CTT TCA GCA TCT TCA CGG-3′, region 2215-2198),12 GDNFR-α (sense, 5′-CGG TTA ACA GCA GGT TGT CAG A-3′, region 669-690; antisense, 5′-GTG TGG GGA TCT CAT TCT CAG AC-3′, region 1469-1447),19 and β-actin (Clontech Laboratories Inc; sense, region 578-609; antisense, region 1415-1384). Primers pairs were selected spanning different introns to distinguish amplified cDNA products from genomic DNA. In the case of RET proto-oncogene, primers amplify a 790-bp region spanning from the extracellular domain upstream the cysteine-rich domain up to the kinase domain downstream the ATP binding site.12 PCR conditions for RET were 4 minutes at 94°C followed by 35 cycles of 45 seconds at 94°C, 45 seconds at 62°C (58°C for GDNFR-α), 60 seconds at 72°C, and a final extension of 5 minutes at 72°C. In the case of β-actin, amplification was performed for 30 cycles according to the manufacturer's guidelines. Fifteen microliters of amplified cDNAs was run in 1.5% agarose gels, blotted onto nylon membranes (Boehringer Mannheim, Mannheim, Germany), and hybridized with 2 × 106 cpm/mL of 5′ end-labeled oligoprobes specifically designated to recognize PCR products. Probes for RET and GDNFR-α spanned nucleotide positions 1430-1454 and 708-735, respectively.12 19
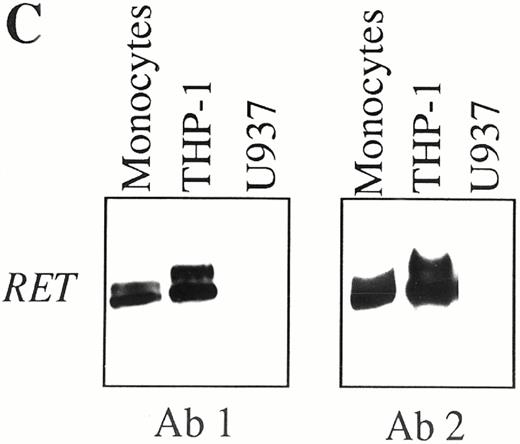
Constitutive expression of RET protein in purified human normal neutrophils, as detected by immunocytochemistry (A and B), and in CD14+ PB monocytes, as detected by Western blotting (C). (A) Neutrophils stained with affinity-purified nonimmune rabbit serum (0.1 μg/mL). (B) Neutrophils stained with affinity-purified rabbit polyclonal antibodies recognizing the RET tyrosine-kinase domain (0.1 μg/mL). Original magnification for (A) and (B) × 400. (C) Western blot analysis of RET protein in human normal monocytes. Proteins extracted from purified CD14+ PB monocytes, THP-1 (positive control), and undifferentiated U937 (negative control) cell lines were immunoprecipitated with two antibodies (Ab 1 and Ab 2) recognizing different cytoplasmic domains the RET RTK, blotted onto immobilon-P membranes, and shown by standard chemiluminescence.
Constitutive expression of RET protein in purified human normal neutrophils, as detected by immunocytochemistry (A and B), and in CD14+ PB monocytes, as detected by Western blotting (C). (A) Neutrophils stained with affinity-purified nonimmune rabbit serum (0.1 μg/mL). (B) Neutrophils stained with affinity-purified rabbit polyclonal antibodies recognizing the RET tyrosine-kinase domain (0.1 μg/mL). Original magnification for (A) and (B) × 400. (C) Western blot analysis of RET protein in human normal monocytes. Proteins extracted from purified CD14+ PB monocytes, THP-1 (positive control), and undifferentiated U937 (negative control) cell lines were immunoprecipitated with two antibodies (Ab 1 and Ab 2) recognizing different cytoplasmic domains the RET RTK, blotted onto immobilon-P membranes, and shown by standard chemiluminescence.
Differences in RET expression in normal and leukemic cells were evaluated by a quantitative and semiquantitative RT-PCR approach. For this purpose, an internal competitor (RETComp ), with a different size than RET-specific amplicons, was prepared from an unrelated DNA fragment engineered to contain specific RET primer templates essentially as described.40,41 Briefly, a BamHI/EcoRI v-erb B 580-bp DNA fragment (Clontech Laboratories Inc) was amplified first with composite primers, containing both RET- and v-erb B-specific sequences, and then with RET gene-specific primers alone. This procedure gave rise to a 597-bp nonhomologous DNA fragment (RETComp ) containing at its ends the appropriate templates for RET primers. RETComp fragments, when amplified with RET-specific primers, yielded to a 597-bp band that was easily discriminated from the 790-bp RET-related amplicon on 1.5% agarose ethidium bromide-stained gels. After purification and densitometric quantitation, a comparison between the amplification kinetics of purified RET amplicons (obtained from THP-1 cells cDNA) and RETComp was performed by amplifying in the same tube 0.1 attomoles of each fragment, removing 5-μL aliquots of reaction mix after 22 to 30 cycles, and analyzing them separately on agarose gels (Fig 1A). A stepwise increase of specific amplified products corresponding to RET (790 bp) and RETComp (597 bp) was observed between 22 and 30 amplification rounds (Fig 1A, upper panel). Quantitation of RET- and RETComp -specific bands by gel analyzer (Gel Doc 1000, BioRad Laboratories, Hercules, CA) resulted in two exponential curves with comparable slopes (Fig 1A, lower panel), indicating a similar amplification efficiency for both fragments. To determine the optimal amount of RETComp for screening studies, 10-fold serial dilutions of RETComp (10 to 1.0 × 10−5 attomoles) were amplified together with constant aliquots (4 μL) of cDNA from the THP-1 cell line, used as a positive control for RET transcripts,28-30 and resolved on agarose gels (Fig 1B, upper panel). Relative intensities of the bands were densitometrically quantitated by computer imaging, expressed as arbitrary units (AU) after correction for the size difference between RET and RETComp , and the logarithms of their ratios were plotted as a function of the logarithm of the amount of RETComp added (Fig 1B, lower panel). This plot was used to determine the equivalence point, ie, the point at which the logarithm of the ratio of RET to RETComp is equal to 0, ie, the amount of RET is equal to the amount of RETComp . In Fig 1B, the equivalence point was inferred between 10−1 and 10−2 attomoles (arrows). Owing to the abundance of RET-specific RNA in the THP-1 cell line,28-30 the use of an RETComp amount about 1 log lower than the equivalence point (ie, 10−3 attomoles) was judged to be optimal for further studies of semiquantitative competitive RT-PCR to detect RET transcripts also in samples with a low expression rate. For such studies, constant amounts of RETComp were amplified by RET-specific primers in the same tube together with 4 μL of reverse-transcribed cDNA from the experimental cell samples in a final 50- μL volume of reaction mix. After resolution on agarose gels, band intensities were quantitated by computer imaging and the relative AU ratios were calculated. Differences in these ratios indicated the relative differences in mRNA levels among the different samples.
Constitutive expression of RET transcripts (semiquantitative RT-PCR) and GDNFR-α (RT-PCR) in human leukemia/lymphoma cell lines. One microgram of total RNA was reverse-transcribed in a 20-μL reaction mix containing hexadeoxyribonucleotides random primers. Conditions for RT-PCR, blotting, hybridization, semiquantitative analysis, and controls were as described in Fig 2.
Constitutive expression of RET transcripts (semiquantitative RT-PCR) and GDNFR-α (RT-PCR) in human leukemia/lymphoma cell lines. One microgram of total RNA was reverse-transcribed in a 20-μL reaction mix containing hexadeoxyribonucleotides random primers. Conditions for RT-PCR, blotting, hybridization, semiquantitative analysis, and controls were as described in Fig 2.
Western blot and immunoprecipitation.Anti-RET antibodies included a rabbit polyclonal antibody raised against the entire RET tyrosine kinase domain (Ab 1) and a rabbit polyclonal antibody raised against a carboxyterminal peptide (residues 1011-1027) of the cytoplasmic domain of human RET (Ab 2),42 both purified by affinity chromatography. Immunoprecipitation and immunoblotting experiments using anti-RET antibodies were performed as previously described.43 Briefly, cells were lysed in 50 mmol/L N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.5, 1% (vol/vol) Triton X-100, 50 mmol/L NaCl, 5 mmol/L EGTA, 50 mmol/L NaF, 20 mmol/L sodium pyrophosphate, 1 mmol/L sodium vanadate, 2 mmol/L phenylsulphonyl fluoride, and 0.2 μg each of aprotinin and leupeptin per milliliter. Lysates were clarified by centrifugation at 10,000g for 15 minutes and the supernatants were processed for immunoprecipitation as described.43 Immunoblots, after incubation with the appropriate antibodies, were shown with the Amersham ECL (Amersham Co, Amersham, UK) system.
Constitutive expression of RET transcripts and GDNFR-α in leukemic cells of myeloid origin (left panels) and in malignant cells from lymphoid tumors (right panels), as assessed by RT-PCR. One microgram of total RNA from AML cases (identified according to their FAB classification) and various lymphoid tumors was reverse-transcribed and amplified with primers specific for RET and GDNFR-α. PCR products were resolved on agarose gel, blotted, and hybridized with specific RET and GDNFR-α oligoprobes. An adult human brain substantia nigra cDNA and cDNA derived from the MN-60 cell line were used, respectively, as positive (+) and negative (−) controls for RET and GDNFR-α expression. cDNAs were always tested with β-actin–specific primers.
Constitutive expression of RET transcripts and GDNFR-α in leukemic cells of myeloid origin (left panels) and in malignant cells from lymphoid tumors (right panels), as assessed by RT-PCR. One microgram of total RNA from AML cases (identified according to their FAB classification) and various lymphoid tumors was reverse-transcribed and amplified with primers specific for RET and GDNFR-α. PCR products were resolved on agarose gel, blotted, and hybridized with specific RET and GDNFR-α oligoprobes. An adult human brain substantia nigra cDNA and cDNA derived from the MN-60 cell line were used, respectively, as positive (+) and negative (−) controls for RET and GDNFR-α expression. cDNAs were always tested with β-actin–specific primers.
Treatment of cells with phosphatidylinositol-specific phospholipase C (PI-PLC) and preparation of a BMSC-derived PI-PLC conditioned medium (PI-PLC/CM).To release GPI-anchored proteins from the cell membrane, primary BMSC, BMF, and the MG-63 osteoblast cell line were preincubated for 1 hour at 37°C with 1 U/mL of PI-PLC19,20 (Boehringer Mannheim), washed three times in IMDM, and then used for flow cytometry analysis (see below). To obtain PI-PLC/CM, 5 to 10 × 106 of primary BMSC were incubated in 1 mL of culture medium in the presence of 1 U/mL of PI-PLC as described above.19 After the removal of cells by centrifugation, the PI-PLC/CM was collected and immediately used for colony assay and flow cytometry.
Leukemic blast colony assay and liquid cultures.The number of leukemic colony-forming units (CFU-L) was assessed as previously described.44 Briefly, 1.0 × 105 T-cell–depleted leukemic blasts were resuspended in 1 mL of IMDM containing 20% FCS and 0.8% methylcellulose and cultured in 100-μL aliquots (6 to 8 replicates) in 96-well flat-bottomed microplates in the presence of increasing concentrations (0.5 to 10 ng/mL) of GDNF (Promega). When indicated, freshly prepared BMSC-derived PI-PLC/CM was added to semisolid medium (10% vol/vol) immediately before plating. After 7 to 14 days of incubation, aggregates with ≥40 cells were scored as colonies.
For differentiation studies, leukemic blast cells (2 × 105/mL) were incubated in the presence of a mixture of GDNF (10 ng/mL) and stromal cell-derived PI-PLC/CM (10% vol/vol), recovered from liquid cultures at different time points, and morphologically analyzed by May-Grünwald-Giemsa staining of cytospin preparation. The percentage of adherent cells was evaluated by scoring the number of cells that needed trypsinization to be detached from the plates compared with the number of cells growing in suspension, as previously described.45 Control experiments were performed by incubating cells either in the presence of GDNF without PI-PLC/CM or with PI-PLC/CM alone.
Immunocytochemistry and flow cytometry.Immunocytochemistry procedures were performed, following the manufacturer's guidelines, with a modified avidin-biotin-complex technique (Kirkegaard & Perry Laboratories Inc, Gaithersburg, MD) on cytospin preparations of purified normal granulocytes, CD14+ monocytes, and selected AML cases. Cytospin slides were fixed in 4% paraformaldehyde plus 0.5% Triton in Tris-HCl buffer and sequentially incubated with affinity-purified rabbit polyclonal antibodies (0.1 μg/mL) recognizing the RET tyrosine-kinase domain,42,43 biotinylated goat antirabbit IgG, and finally alkaline phosphatase-conjugated streptavidin. Immunostaining was developed by incubating the slides with the appropriate chromogenic substrates (Kirkegaard & Perry Laboratories Inc) followed by hematoxylin (Sigma) counterstain. Controls included the omission of the primary antibody and the use of affinity-purified nonimmune rabbit serum (0.1 μg/mL). For flow cytometric detection of surface GPI-linked GDNF receptors (GDNFR-α),20 BMSC, BMF, and MG-63 cells, pretreated or not with PI-PLC as described above, were exposed for 1 hour at 37°C with GDNF (100 ng/mL; Promega) and then incubated sequentially with chicken polyclonal antihuman GDNF (100 μg/mL; Promega) and rabbit fluorescein isothiocyanate-conjugated isotype-matched antichicken Igs (Promega). As controls, isotype-matched irrelevant chicken Igs (Promega) were used. In some experiments, primary leukemic cells were exposed for 1 hour at 37°C to GDNF alone (100 μg/mL) or to a combination of GDNF and BMSC-derived PI-PLC/CM, before being processed for flow cytometric detection of membrane-bound GDNF. Viable, antibody-labeled cells were identified according to their forward and side scatter, electronically gated, and assayed for surface fluorescence on a FACScan flow cytometer (Becton Dickinson, Mountain View, CA).
RESULTS
Expression of RET proto-oncogene and GDNFR-α by normal hematolymphoid cells.cDNAs obtained from purified cell populations were assayed for RET expression by semiquantitative RT-PCR and the specificity of amplified products was confirmed by Southern blotting hybridization with an internal RET oligoprobe. As shown in Fig 2, a faint 790-bp band corresponding to RET amplified products was detected in CD34+ hematopoietic progenitors from mobilized PB and cord blood (Fig 2). As compared with early progenitors, higher levels of RET transcripts were found in circulating neutrophils (5-fold) and adherent CD14+ monocyte/macrophages (6-fold), being further increased (11-fold) in these latter cells upon GM-CSF–induced cellular activation (Fig 2). In contrast, circulating eosinophils and different T-cell subsets from PB, thymus, and tonsils did not express RET-specific mRNA, which remained undetectable in normal peripheral T cells also after cellular activation by TPA and ionomycin (Fig 2). Similarly, tonsil and PB CD19+ B cells were negative for RET mRNA (Fig 2). As opposed to RET proto-oncogene, transcripts specific for GDNFR-α were never found in all the normal cell types analyzed (Fig 2).
In agreement with RT-PCR data, expression of RET protein was detected by immunocytochemistry and Western blotting both in purified neutrophils and in CD14+ peripheral monocytes (Fig 3 and data not shown). In the case of neutrophils, a prominent granular staining of cytoplasms, along with a faint membrane reactivity in some of the cells, was usually observed (Fig 3B), whereas in monocytes a strong and diffuse cytoplasmic staining was commonly associated with a clear membrane labeling (data not shown). As shown in Fig 3C, RET products of 150 and 170 kD were detected in CD14+ purified monocytes by Western blotting with two different anti-RET polyclonal antibodies (Ab 1 and Ab 2). In agreement with previous data,30RET-specific components of 150 and 190 kD were found with both antibodies in the THP-1 cell line, which was used as a positive control, whereas no RET protein was immunodetected in undifferentiated U937 cells (Fig 3C).
Expression of RET proto-oncogene and GDNFR-α by human leukemic cell lines of myeloid and lymphoid lineages.By studying a large panel of human leukemia/lymphoma cell lines, RET mRNA was detected, albeit at different constitutive levels, in malignant cells of both myeloid and lymphoid origin. As shown in Table 1 and Fig 4, the highest relative levels of RET transcripts were detected in cell lines of the myelomonocytic lineage, whereas most tumor B-cell lines, encompassing early pre-B to plasmacell differentiation stages, were either negative (11/13) or displayed (Nalm-6 and IM-9) low levels of RET mRNA. Among cell lines of myelo-granulocytic (KG-1, KG-1A, HL-60, and NB-4) and monocytic (U937, ML-3, and THP-1) phenotypes, a correlation between the constitutive expression of RET proto-oncogene and the relative maturation stage was observed (Fig 4). In particular, the early (CD34+) myeloblasts cell line KG-1A displayed low levels of RET mRNA, which were more than threefolds and greater than fivefold higher in the intermediate myeloblast cell line HL-60 and in the promyelocytic leukemia cell line NB-4, respectively (Fig 4). Similarly, the highest amount of RET transcripts among monocytic cell lines was detected in those (THP-1) displaying the more mature phenotype,34 as compared with ML-3 and undifferentiated U937 (Fig 4). Accordingly, a significant upregulation of RET mRNA levels was observed by semiquantitative RT-PCR upon induction of monocytic maturation of U937 cells by either TPA (8-fold at day 3) and vitamin D3 plus TGF-β (23-fold at day 6), along with a parallel increase of immunodetectable RET protein (data not shown). No RET expression was detected in cell lines of erythroid derivation (K562 and HEL) and osteoclast phenotype (FLG 29.1), whereas very low levels of RET transcripts were found in the megakaryocytic cell line Mo7e (Fig 4). Although the relative amount of RET transcripts detected in human leukemia/lymphoma T-cell lines was overall lower than in myeloid cell lines, a correlation between the levels of RET expression and the relative maturation stage again emerged from our analysis (Fig 4). As shown by semiquantitative RT-PCR assay, cell lines of mature T-cell phenotype (H9, HUT 78, and Karpas-299) displayed RET mRNA levels about fivefold higher than cell lines of postthymic phenotype (Jurkat and KE-37), whereas no expression of the proto-oncogene was found in cell lines (Molt-4 and FRO) of thymic phenotype (Table 1 and Fig 4). As compared with RET, the expression of GDNFR-α was very infrequent, being confined to isolated cell lines of myelomonocytic cell (KG-1A), T-cell (Molt-4), and B-cell (Nalm-6) phenotypes (Table 1 and Fig 4).
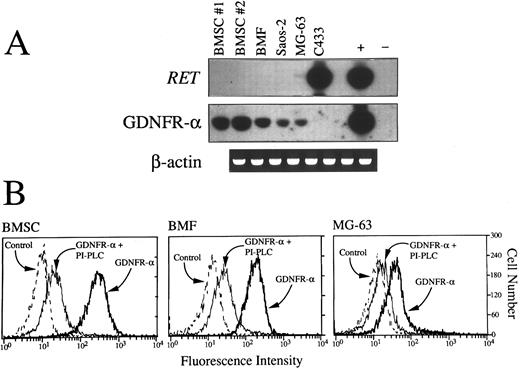
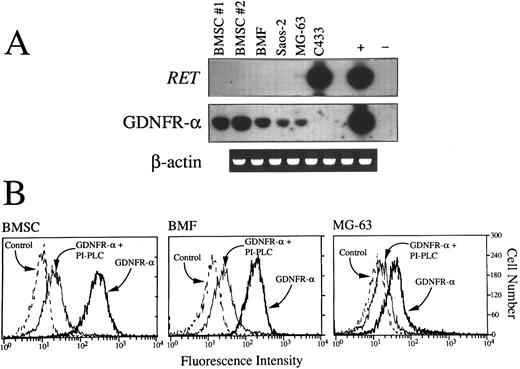
(A) RT-PCR detection of RET and GDNFR-α mRNA in primary BMSC from two donors (nos. 1 and 2), BMF, Saos-2, and MG-63 osteoblast cell lines and C433 stromal cell line. Aliquots of cDNA bulks were amplified with primer pairs specific for RET and GDNFR-α, run in agarose gels, blotted, and hybridized with oligoprobes specifically designated to recognize PCR-amplified products. An adult human brain substantia nigra cDNA and cDNA derived from the MN-60 cell line were used, respectively, as positive (+) and negative (−) controls for RET and GDNFR-α expression. cDNAs were always tested with β-actin–specific primers. (B) Fluorescence histograms showing expression of surface GPI-linked GDNF receptors (GDNFR-α) in BMSC, BMF, and the MG-63 cell line. Cells pretreated or not with PI-PLC (1 U/mL for 1 hour at 37°C) were incubated for 1 additional hour with GDNF (100 ng/mL) and then sequentially with chicken polyclonal antihuman GDNF (100 μg/mL) and rabbit fluorescein isothiocyanate-conjugated isotype-matched antichicken Igs. As controls, isotype-matched irrelevant chicken Igs were used.
(A) RT-PCR detection of RET and GDNFR-α mRNA in primary BMSC from two donors (nos. 1 and 2), BMF, Saos-2, and MG-63 osteoblast cell lines and C433 stromal cell line. Aliquots of cDNA bulks were amplified with primer pairs specific for RET and GDNFR-α, run in agarose gels, blotted, and hybridized with oligoprobes specifically designated to recognize PCR-amplified products. An adult human brain substantia nigra cDNA and cDNA derived from the MN-60 cell line were used, respectively, as positive (+) and negative (−) controls for RET and GDNFR-α expression. cDNAs were always tested with β-actin–specific primers. (B) Fluorescence histograms showing expression of surface GPI-linked GDNF receptors (GDNFR-α) in BMSC, BMF, and the MG-63 cell line. Cells pretreated or not with PI-PLC (1 U/mL for 1 hour at 37°C) were incubated for 1 additional hour with GDNF (100 ng/mL) and then sequentially with chicken polyclonal antihuman GDNF (100 μg/mL) and rabbit fluorescein isothiocyanate-conjugated isotype-matched antichicken Igs. As controls, isotype-matched irrelevant chicken Igs were used.
Expression of RET proto-oncogene and GDNFR-α by primary leukemic cells of myeloid or lymphoid lineages.A broad expression of the RET proto-oncogene was found in AML, with about 60% of cases (32/53) displaying significant amounts of RET RNA (Table 2 and Fig 5). In particular, a very high frequence of RET expression (17/24 [71%]) was detected among the monocytic and myelomonocytic subtypes of AML (FAB M4 and M5) as compared with immature myeloid phenotypes (FAB M0 and M1), in which RET-positive samples accounted for 31% (5/16) of cases (Table 2 and Fig 5). In addition, almost all cases (6/7) of acute promyelocytic leukemia (FAB M3) and FAB M2 AMLs (3/4) displayed RET transcripts, which were also detected in 3 of 5 samples of MBC (Table 2). Accordingly, expression of RET protein was detected by immunocytochemistry of blast cells from selected RET-expressing AML cases (2 FAB-M3, 5 FAB-M4, and 3 FAB-M5), showing a strong cytoplasmic staining usually associated with a clear membrane labeling in monocytic-oriented cytotypes (data not shown). Among neoplasms of lymphoid origin (Table 3 and Fig 5), RET expression was a very infrequent event, being detected in 2 of 9 cases of T-ALLs, in a single case of ATLL (CD4+/CD8−), in 2 of 7 samples of B-precursor ALL, and in scattered cases of B-CLL (5/45), HCL (1/7), high-grade (2/9) or low-grade (2/9) NHL in overt leukemic phase, and MM (1/6).
As seen in normal hematopoietic cells and leukemic cell lines, the broad expression of RET proto-oncogene in primary tumor cells was not paralleled by a mutual expression of GDNFR-α mRNA. In our series, only two isolated cases (1 AML and 1 T-PLL) were found to express GDNFR-α–specific transcripts (Tables 2 and 3 and Fig 5).
Expression of RET proto-oncogene and GDNFR-α by BM stromal cells.The broad expression of RET tyrosine kinase by normal and malignant hematopoietic cells in the absence of the naturally occurring receptor for GDNF prompted us to look for other possible sources of GDNFR-α within the accessory cells of the hematopoietic microenvironment. As shown in Fig 6A and B, a high amount of GDNFR-α transcripts and GDNF-binding sites was found in normal BMSC from two different donors (nos. 1 and 2 in Fig 6A), BMF, and, albeit at a lower level, two osteoblast cell lines (Saos-2 and MG-63) previously described to support normal hematopoiesis.46 The binding of GDNF to stromal cells was virtually abolished after treatment of cells with PI-PLC (Fig 6B), which specifically removes GPI-linked molecules from the cell surface. Interestingly, the expression of GDNFR-α mRNA in primary stromal cells and stromal cell lines was not associated with the presence of RET, which was conversely strongly expressed in the absence of GDNFR-α transcripts by another cell line (C433)9 derived from the stromal component of an osteoclastoma (Fig 6A).
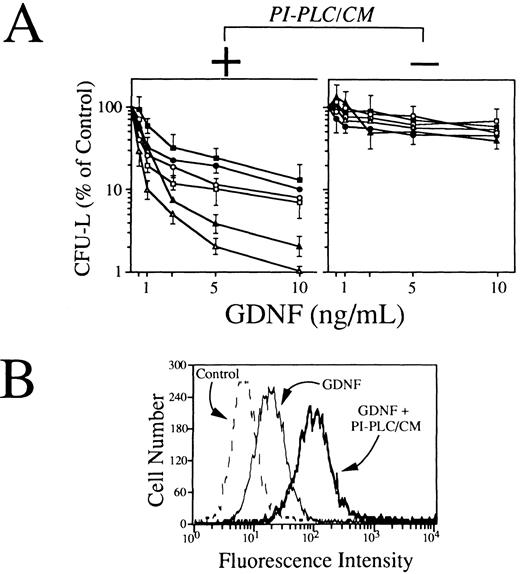
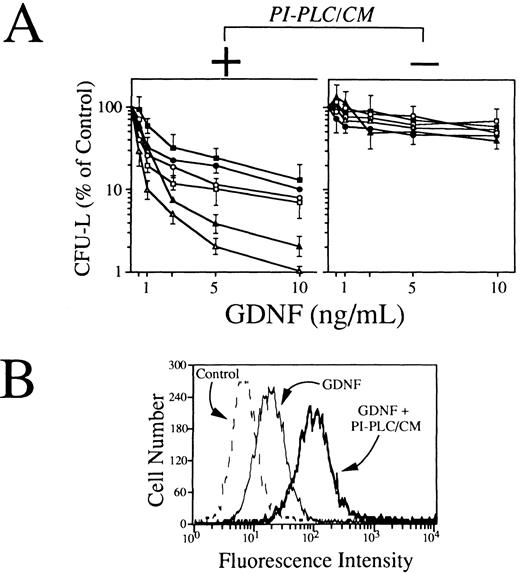
Effects of GDNF on clonogenic growth and monocytic differentiation of human leukemic cells.Because RET-expressing AML cells do not produce the ligand binding component for GDNF, we have performed functional experiments by using conditioned media from stromal cells as a putative source of soluble GDNFR-α and/or other GPI-linked GDNF receptors. To this end, T-lymphocyte–depleted mononuclear cells from 6 RET-positive AMLs of myelomonocytic phenotype (FAB M4 and M5) were exposed at various concentrations of GDNF (0.5 to 10 ng/mL) in a standard clonogenic assay in the presence or not of a fixed concentration (10% vol/vol) of BMSC-derived PI-PLC/CM. As shown in Fig 7A (left panel), GDNF, when associated to BMSC-derived PI-PLC/CM, induced a dose-dependent impairment of leukemic blasts clonogenic growth, yielding more than 80% to 90% inhibition of CFU-L at a concentration of 10 ng/mL (Fig 6A). Conversely, in the absence of BMSC-derived PI-PLC/CM, GDNF-mediated effects of leukemic cell growth were strikingly reduced (Fig 6A, right panel). The inhibitory effects exerted by GDNF alone on RET-expressing leukemic cells were overall comparable to those observed in 3 cases of RET-negative AML cells grown either in the presence or not of BMSC-derived PI-PLC/CM (data not shown). The ability of GDNF to bind RET-expressing primary leukemic cells was assessed by a flow cytometric staining with polyclonal anti-GDNF antibodies in the presence or not of BMSC-derived PI-PLC/CM. Our results indicated that, in agreement with clonogenic data, GDNF was able to efficiently bind leukemic cells in the precence of BMSC-derived PI-PLC/CM (Fig 7B). GDNF binding to primary leukemic cells was not increased in the presence of PI-PLC/CM derived from cells not expressing GDNFR-α mRNA and used as internal negative control for these experiments (data not shown), suggesting that GDNFR-α could actually represent the major GDNF-binding activity produced by stromal cells.
(A) Effects of GDNF alone (right panel) or in association with a fixed concentration (10% vol/vol) of conditioned medium derived from stromal cells exposed to PI-PLC (PI-PLC/CM; left panel) on clonogenic growth of blast cells derived from 6 RET-expressing AML samples. T-cell–depleted leukemic blasts (1.0 × 105) were resuspended in 1 mL of IMDM containing 20% FCS and 0.8% methylcellulose and cultured in 100-μL aliquots in 96-well flat-bottomed microplates in the presence of increasing concentrations (0.5 to 10 ng/mL) of GDNF. Freshly prepared BMSC-derived PI-PLC/CM was added to semisolid medium (10% vol/vol) immediately before plating. After 7 days of incubation, aggregates with ≥40 cells were scored as colonies. Results are expressed as mean ± SEM of 6 to 8 replicates. (B) Fluorescence histograms showing the expression of membrane-associated GDNF/soluble GDNF receptors complexes by primary RET-expressing AML cells. Cells were exposed for 1 hour at 37°C to GDNF alone (100 μg/mL) or to a combination of GDNF and BMSC-derived PI-PLC/CM and then sequentially with chicken polyclonal antihuman GDNF (100 μg/mL) and rabbit fluorescein isothiocyanate-conjugated isotype-matched antichicken Igs. As controls, isotype-matched irrelevant chicken Igs were used.
(A) Effects of GDNF alone (right panel) or in association with a fixed concentration (10% vol/vol) of conditioned medium derived from stromal cells exposed to PI-PLC (PI-PLC/CM; left panel) on clonogenic growth of blast cells derived from 6 RET-expressing AML samples. T-cell–depleted leukemic blasts (1.0 × 105) were resuspended in 1 mL of IMDM containing 20% FCS and 0.8% methylcellulose and cultured in 100-μL aliquots in 96-well flat-bottomed microplates in the presence of increasing concentrations (0.5 to 10 ng/mL) of GDNF. Freshly prepared BMSC-derived PI-PLC/CM was added to semisolid medium (10% vol/vol) immediately before plating. After 7 days of incubation, aggregates with ≥40 cells were scored as colonies. Results are expressed as mean ± SEM of 6 to 8 replicates. (B) Fluorescence histograms showing the expression of membrane-associated GDNF/soluble GDNF receptors complexes by primary RET-expressing AML cells. Cells were exposed for 1 hour at 37°C to GDNF alone (100 μg/mL) or to a combination of GDNF and BMSC-derived PI-PLC/CM and then sequentially with chicken polyclonal antihuman GDNF (100 μg/mL) and rabbit fluorescein isothiocyanate-conjugated isotype-matched antichicken Igs. As controls, isotype-matched irrelevant chicken Igs were used.
To better clarify mechanisms underlying GDNF growth inhibitory effects on primary AML cells, T-cell–depleted RET-expressing leukemic blasts (2 AML FAB M4 and 1 AML FAB M5) were exposed in liquid culture to 10 ng/mL of GDNF in the presence or absence of BMSC-derived PI-PLC/CM. As shown in Fig 8, the concurrent exposure of leukemic blasts to GDNF and BMSC-derived PI-PLC/CM resulted in a time-dependent increase in the number of cells adhering to the plastic (Fig 8A and B) and displaying, after 5 days of culture, morphologic changes consistent with differentiation towards mature monocyte/macrophages (Fig 8C and D). Exposure of blast cells to GDNF alone or BMSC-derived PI-PLC/CM alone did not induce significant changes in the adherence (Fig 8A) and the morphologic appearance of leukemic cells (not shown). An increased expression of CD14, CD11b, and CD15 antigens was also found in leukemic cells after exposure to GDNF and BMSC-derived PI-PLC/CM (not shown).
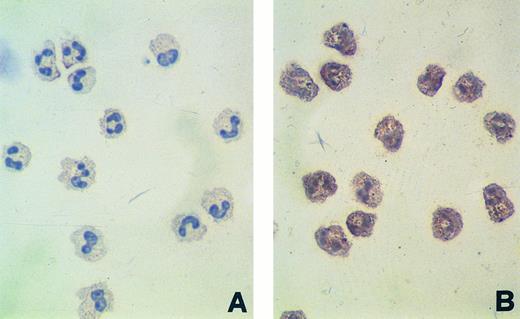
(A) Adherence of leukemic cells from 3 RET-expressing AML samples cultured in medium alone (open symbols) or in the presence of a mixture of GDNF (10 ng/mL) and BMSC-derived PI-PLC/CM (10% vol/vol) (solid symbols). As further controls, the percentage of adherent cells was also evaluated in cultures performed for 5 days in the presence of GDNF alone (solid symbols at the far right) and BMSC-derived PI-PLC/CM alone (open symbols at the far right). The percentage of cells adhering to plastic dishes was was evaluated by scoring the number of cells that needed trypsinization to be detached from plates, compared with the number of cells growing in suspension. Results are mean ± SEM of triplicate cultures. (B) Phase-contrast micrograph of differentiated blasts from a 5-day-old culture performed in the presence of GDNF (10 ng/mL) and BMSC-derived PI-PLC/CM (10% vol/vol) (original magnification × 200). (C and D) Morphologic appearance of leukemic blasts from a RET-expressing AML (FAB-M5) sample cultured for 5 days in the absence (C) or in the presence of GDNF (10 ng/mL) and BMSC-derived PI-PLC/CM (10% vol/vol) (D); May-Grünwald-Giemsa staining of cytospin preparation (original magnification for [C] and [D] × 630).
(A) Adherence of leukemic cells from 3 RET-expressing AML samples cultured in medium alone (open symbols) or in the presence of a mixture of GDNF (10 ng/mL) and BMSC-derived PI-PLC/CM (10% vol/vol) (solid symbols). As further controls, the percentage of adherent cells was also evaluated in cultures performed for 5 days in the presence of GDNF alone (solid symbols at the far right) and BMSC-derived PI-PLC/CM alone (open symbols at the far right). The percentage of cells adhering to plastic dishes was was evaluated by scoring the number of cells that needed trypsinization to be detached from plates, compared with the number of cells growing in suspension. Results are mean ± SEM of triplicate cultures. (B) Phase-contrast micrograph of differentiated blasts from a 5-day-old culture performed in the presence of GDNF (10 ng/mL) and BMSC-derived PI-PLC/CM (10% vol/vol) (original magnification × 200). (C and D) Morphologic appearance of leukemic blasts from a RET-expressing AML (FAB-M5) sample cultured for 5 days in the absence (C) or in the presence of GDNF (10 ng/mL) and BMSC-derived PI-PLC/CM (10% vol/vol) (D); May-Grünwald-Giemsa staining of cytospin preparation (original magnification for [C] and [D] × 630).
DISCUSSION
A putative role of the RET-encoded RTK in the functional regulation of hematopoietic cells was suggested by the presence of RET transcripts and protein in lympho-hematopoietic tissues of mice and rats, including fetal liver, thymus, spleen, and lymph nodes,22,47,48 and in two human leukemic cell lines (HL-60 and THP-1).28-30 Despite these preliminary indications, expression of the RET RTK in human lymphohematopoietic cells has not been investigated in detail so far.
In the present study, we have shown that relatively low amounts of RET mRNA can be found in early CD34+ hematopoietic progenitors, but RET transcripts appeared to increase with maturation along the myelomonocytic lineage, being upregulated in circulating neutrophils and resting or activated monocytes. These results were confirmed at a protein level by immunostaining and Western blotting. Analysis of human myelomonocytic cell lines was consistent overall with the pattern of normal cells, showing a progressive increase of RET transcripts in cells representing early to late stages of granulocytic and monocytic differentiation,34 being further upregulated during in vitro maturation of monoblastic cell lines. Accordingly, we have shown that RET expression is mainly confined to AML displaying either a monocytic (FAB M4 and M5) or an intermediate-mature myeloid (M2 and M3) phenotype, being less frequently detected in early myeloid (M0 and M1) AMLs. Taken together, our results indicate that RET expression is maturation-associated in human myelopoiesis, suggesting a possible role of RET product in the functional regulation of intermediate and mature myelomonocytic cells. In this regard, RET behavior appeared to diverge from that of most hematopoietic TRKs, including KIT,FLT3,TIE, and TNK1, which are usually expressed at a very high level in early CD34+ progenitors and downregulated or switched off after maturation towards granulocytes and/or monocytes.2,10 49-51
In contrast, by analyzing normal and malignant B cells encompassing different stages of B-cell maturation, RET transcripts were only sporadically detected in SIg+ B-cell tumors (5/72). These results are in overall agreement with data from Wasserman et al52 showing that, in mouse B cells, RET is expressed only in early stages of B-cell differentiation, being drastically downregulated after the expression of surface Igs. However, we were not able to detect RET mRNA in most early B-lineage ALLs of pro-B and pre-B phenotypes. The discrepant behavior of RET in the early steps of mouse and human B-cell lymphopoiesis closely parallels that of the KIT-encoded RTK, which, although functionally expressed in mouse early B cells (from pro-B to late pre-B stage),53,54 is not usually detectable in human B-lineage ALLs.55
As seen for B cells, purified normal T cells from PB, tonsil, and thymus never expressed RET proto-oncogene, even after cellular activation. However, a significant amount of RET mRNA was detected in some tumor cells of postthymic and mature T-cell phenotype, most of them derived from dermatotropic T-cell malignancies, including cutaneous T-cell lymphomas (H9, HUT 78),35 CD30+ anaplastic large-cell lymphoma (Karpas-299),56 and ATLL.57 One can therefore speculate that the expression of RET RTK may be somehow associated with cell types characterized by a high migratory ability, such as neural crest elements,21,22,28,47,48 58 and, as shown here, monocytes, neutrophils, and dermatotropic malignant T cells.
The expression of the RET gene in normal and malignant hematopoietic cells raises the question of the functional significance of this RTK in the human hematopoietic compartment,59 in addition to its previously defined role in the regulation of developing central and peripheral nervous system and kidney.21-25 It has been recently shown that RET RTK is involved in the formation at the cell surface of a complex receptor system for GDNF,15,16 in which a GPI-linked molecule (GDNFR-α) acts as a ligand-binding component and RET represents the signaling component.19,20 It appears therefore that RET-expressing cells may transduce GDNF-mediated signals in the presence of GDNFR-α.19,20 However, by analyzing a large variety of purified normal cells, leukemic cell lines, and primary leukemia/lymphoma cells of myeloid and lymphoid origin, we were unable to show a consistent expression of GDNFR-α in human hematopoietic cells, irrespective of the presence of the RET RTK. Because GDNFR-α is an extracellular protein that is attached to the outer cell membrane19,20 and RET activation can be induced by GDNF in cells not expressing GDNFR-α in the presence of culture media containing soluble GDNFR-α,19,20 we speculated that accessory cells of the BM microenvironment could provide a physiologic source of GDNFR-α for RET-expressing hematopoietic cells. In agreement with such a view, we were able to show that human BMSC, BMF, and other stromal cell lines capable of supporting normal hematopoiesis6,46 do not express RET but produce high levels of GDNFR-α mRNA and surface GPI-linked GDNF receptors, most probably including GDNFR-α. Accordingly, we have provided evidence that RET-expressing human AML cells are able to efficiently bind exogenous GDNF in the presence of supernatants derived from stromal cells treated with PI-PLC to remove GPI-linked proteins (PI-PLC/CM) and that GDNF reduces the clonogenic capacity of leukemic cells in the presence of stromal cell-derived PI-PLC/CM. In addition, we have shown that PI-PLC/CM from cells not producing GDNFR-α did not increase binding and biologic effects of GDNF on RET-expressing leukemic cells and that clonogenic ability of RET-negative AML cells is not significantly modified by GDNF, also in the presence of stromal cell-derived PI-PLC/CM. These data overall support the idea that GDNFR-α or a similar, yet unidentified, GPI-linked receptor for GDNF produced by cells of the BM microenvironment can mediate the action of this neutrotrophin on human leukemic cells. The mechanisms underlying the inhibitory effects of GDNF on human AML cells are still obscure, but our present results seem to indicate that GDNF may induce terminal division and differentiation of monocytic leukemia cells. Accordingly, we have obtained data that GDNF is able to enhance both the generation and maturation of monocyte/macrophage precursors (CFU-M) from normal BM CD34+ cells in the presence of stromal cell-derived PI-PLC/CM (Gattei et al, manuscript in preparation). Such a possibility is in agreement with the previous demonstration that GDNF is able to induce morphologic and functional differentiation of neural and developing renal cells.17,23-25 The involvement of GDNF in the functional regulation of normal and neoplastic monocytic cells is also supported by the close developmental relationships between glial cells and the monocyte/macrophage system60 and by the high levels of expression of this neurotrophic factor in nonneural tissues, including hematopoietic organs such as liver and spleen.19,20 61
In light of the emerging role of neurotrophins in the regulation of hematopoietic and immune cells62,63 and of the documented involvement of the RET RTK in a number of neoplastic and nonneoplastic human diseases,26,27,42 58 we are currently investigating the relationships between accessory cell-derived GDNFR-α–, GDNF-, and RET-expressing cells within the normal and neoplastic lymphohematopoietic microenvironment.
ACKNOWLEDGMENT
We are grateful to Dr N. Dathan for providing RET antibodies and to Dr L. De Marco (Blood Transfusion Center, C.R.O. Aviano, Aviano, Italy) for kindly providing PB apheresis products. We also gratefully acknowledge the excellent assistance of Fulvio Coletto for the artwork and graphic support.
A. Celetti and A. Cerrato equally contributed to this work.
Supported by the Associazione Italiana per la Ricerca sul Cancro (Milano, Italy); the Ministero della Università, Ricerca Scientifica e Tecnologica (Rome, Italy); the Consiglio Nazionale delle Ricerche, PF-ACRO, Italy; and the Ministero della Sanità, Ricerca Finalizzata IRCCS (Rome, Italy). Part of this work was performed while G.V. was holding a position of Fogarty-Scholar-in-Residence at the National Institutes of Health, Bethesda, MD.
Address reprint requests to Michele Grieco, MD, Dipartimento di Medicina Sperimentale e Clinica, Facoltà di Medicina e Chirurgia, Università degli Studi di Reggio Calabria, Via T. Campanella, I-88100, Catanzaro, Italy.






![Fig. 8. (A) Adherence of leukemic cells from 3 RET-expressing AML samples cultured in medium alone (open symbols) or in the presence of a mixture of GDNF (10 ng/mL) and BMSC-derived PI-PLC/CM (10% vol/vol) (solid symbols). As further controls, the percentage of adherent cells was also evaluated in cultures performed for 5 days in the presence of GDNF alone (solid symbols at the far right) and BMSC-derived PI-PLC/CM alone (open symbols at the far right). The percentage of cells adhering to plastic dishes was was evaluated by scoring the number of cells that needed trypsinization to be detached from plates, compared with the number of cells growing in suspension. Results are mean ± SEM of triplicate cultures. (B) Phase-contrast micrograph of differentiated blasts from a 5-day-old culture performed in the presence of GDNF (10 ng/mL) and BMSC-derived PI-PLC/CM (10% vol/vol) (original magnification × 200). (C and D) Morphologic appearance of leukemic blasts from a RET-expressing AML (FAB-M5) sample cultured for 5 days in the absence (C) or in the presence of GDNF (10 ng/mL) and BMSC-derived PI-PLC/CM (10% vol/vol) (D); May-Grünwald-Giemsa staining of cytospin preparation (original magnification for [C] and [D] × 630).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/8/10.1182_blood.v89.8.2925/4/m_bl_0037f8.jpeg?Expires=1765954411&Signature=hzLfsYBpvQYGnjLNa5yU2Dqaa7NzrOfyMdkQNHJFP0MJ0hTuykuiLqn-HTxqns6X2dQhVi6n14KCQmsEr6sXNGOO-T1b5pFksQEG8r~fU2x4Axxpf9Wh3f4wwTCwyHjIVzLicgoUq--zZxRbTFkeJkxWriaLqgcCp5Nuf75tpOhXNmU8PdcSjHbLORz5QxZIAbk4Us0urg-EuKWR-3A1~k4hJvsIBmsQ9nxLFg5Od4HHPtUAG2HCPc8D5j6Y-39eZEYoP1wnd2M0~lEw2oMN4j-TUtWA~2GXgkgR--TG7dH28lYPC6RCE1aNry1vbXW-nN6vNWJmdCYQjo~k83nqwg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





![Fig. 8. (A) Adherence of leukemic cells from 3 RET-expressing AML samples cultured in medium alone (open symbols) or in the presence of a mixture of GDNF (10 ng/mL) and BMSC-derived PI-PLC/CM (10% vol/vol) (solid symbols). As further controls, the percentage of adherent cells was also evaluated in cultures performed for 5 days in the presence of GDNF alone (solid symbols at the far right) and BMSC-derived PI-PLC/CM alone (open symbols at the far right). The percentage of cells adhering to plastic dishes was was evaluated by scoring the number of cells that needed trypsinization to be detached from plates, compared with the number of cells growing in suspension. Results are mean ± SEM of triplicate cultures. (B) Phase-contrast micrograph of differentiated blasts from a 5-day-old culture performed in the presence of GDNF (10 ng/mL) and BMSC-derived PI-PLC/CM (10% vol/vol) (original magnification × 200). (C and D) Morphologic appearance of leukemic blasts from a RET-expressing AML (FAB-M5) sample cultured for 5 days in the absence (C) or in the presence of GDNF (10 ng/mL) and BMSC-derived PI-PLC/CM (10% vol/vol) (D); May-Grünwald-Giemsa staining of cytospin preparation (original magnification for [C] and [D] × 630).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/8/10.1182_blood.v89.8.2925/4/m_bl_0037f8.jpeg?Expires=1765954412&Signature=Hvgc5QyCRgHKgyqHtc1yod~bxcuJ7j2PoGtK1PkFvwN-MLeIwySsNnYtOsA82S5NO0BYHuT681GeVifz9DJoZBr~PGweKphExxoS0clYo4WNl~oP5T9gMsioymm4LXm0EQmQjTDsMSayhG1TfNxd69NlzeUZZW~PqyzOwVcB8~IGO6obgdh9Nv4p4lL8-4rQ75YeitTgX4x4VygM12la35D88WcTsn7VJaQvKRuMrDpDr9YqB2on0kgSPdplRM5tGRmbTlGGCSU3XMieec9Lk5Ln4f3-GW2GgHYw6f6PLkj2e9NZOy~6xrMV8qTOIcPsAveMTSrCLKfebcCTvu8JEg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)