Abstract
Activated neutrophils have the ability to upregulate the expression of many genes, in particular those encoding cytokines and chemokines, and to subsequently release the corresponding proteins. Although little is known to date concerning the regulation of gene transcription in neutrophils, it is noteworthy that many of these genes depend on the activation of transcription factors, such as NF-κB, for inducible expression. We therefore investigated whether NF-κB/Rel proteins are expressed in human neutrophils, as well as their fate on cell activation. We now report that dimers consisting of p50 NFκB1, p65 RelA, and/or c-Rel are present in neutrophils and that the greater part of these protein complexes is physically associated with cytoplasmic IκB-α in resting cells. Following neutrophil stimulation with proinflammatory agonists (such as lipopolysaccharide [LPS], tumor necrosis factor-α [TNF-α], and fMet-Leu-Phe) that induce the production of cytokines and chemokines in these cells, NF-κB/Rel proteins translocated to nuclear fractions, resulting in a transient induction of NF-κB DNA binding activity, as determined in gel mobility shift assays. The onset of both processes was found to be closely paralleled by, and dependent on, IκB-α degradation. Proinflammatory neutrophil stimuli also promoted the accumulation of IκB-α mRNA transcripts, resulting in the reexpression of the IκB-α protein. To our knowledge, this constitutes the first indication that NF-κB activation may underlie the action of proinflammatory stimuli towards human neutrophil gene expression and, as such, adds a new facet to our understanding of neutrophil biology.
NEUTROPHILS RANK among the first blood cells that migrate towards inflammatory lesions, where they accumulate in large numbers and perform host defense functions. These include the phagocytosis of invading microorganisms or of foreign particles, the release of proteolytic enzymes, the generation of oxygen-derived reactive intermediates, and the synthesis of potent lipid mediators such as leukotriene B4 and platelet-activating factor (PAF ). As a result, the relevance of neutrophils to host immunity has traditionally been viewed as being restricted to these effector functions. In recent years, however, it has become evident that under various stimulatory conditions, neutrophils also have the ability to synthesize and release many proteins that can influence the course of inflammatory and immune responses. A partial list of such proteins includes cytokines and related products, such as tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), and the IL-1 receptor antagonist (IL-1ra), as well as chemokines such as IL-8, macrophage inflammatory protein-1 α (MIP-1α), and MIP-1β.1 Therefore, these observations emphasize the pertinence of considering neutrophils not only as professional phagocytes, but also as cells that can play a pivotal role in orchestrating inflammatory and immune processes.
The inducible production of these immunoregulatory proteins by neutrophils is usually preceded by, and largely dependent on, an accumulation of the corresponding mRNA transcripts,2-8 a process that is generally prevented by pretreating the cells with the transcription inhibitor, actinomycin D5 (and our unpublished observations, May 1992). It follows that transcriptional events are likely to play a central role in the induction of cytokine and chemokine production in neutrophils.9,10 Although little is known to date concerning the regulation of gene transcription in neutrophils, a number of clues point to the potential involvement of distinct transcription factors in this process. First, a common characteristic of the genes encoding TNF-α, IL-1β, IL-1ra, IL-8, and MIP-1α, is that they all feature κB or κB-like binding motifs in their 5′ regulatory region.11-16 Moreover, the binding of NF-κB dimers to these motifs either suffices to confer transcriptional inducibility,12-15 or in the case of the IL-1ra gene, is required for full promoter activation.16 Finally, a markedly increased expression of the transcripts encoding the above cytokines and chemokines can be induced in neutrophils by stimuli such as lipopolysaccharide (LPS) or TNF-α,2-8,17 which are known to be potent activators of NF-κB in other systems.18 19
The transcription factor, NF-κB, consists of homo- or heterodimers of the Rel family proteins, p50/NFκB1, p52/NFκB2, p65/RelA, and c-Rel. In most cell types studied to date, NF-κB dimers are retained in the cytoplasm through a physical association with inhibitor proteins, termed IκB.20 Following cell activation, IκB becomes hyperphosphorylated on distinct serine residues, and a mounting body of evidence indicates that this hyperphosphorylation targets the inhibitor for proteolytic degradation (reviewed in Finco and Baldwin21 ). The degradation of IκB eventually leads to its dissociation from NF-κB dimers, thereby allowing the movement of the latter towards the nucleus, where they may bind with high specificity to enhancer sequences in the 5′ regulatory region of target genes. Among those genes whose expression is most rapidly enhanced in a κB-dependent manner is the one that encodes IκB-α.22-25 This, in turn, leads to the rapid resynthesis of the inhibitor protein,22,23 of which a portion accumulates in the nucleus where it is able to dissociate NF-κB dimers from DNA.26 Fragmentary evidence also points to a potential role for IκB-α in retargeting NF-κB dimers from the nucleus to the cytoplasm,26 27 but the mechanism whereby this may occur remains unknown.
In the current study, we investigated the expression and distribution of NF-κB/Rel proteins and their cytoplasmic inhibitor, IκB-α, in human peripheral blood neutrophils, as well as their respective fate on cell activation. We now report that following stimulation of neutrophils with proinflammatory mediators that are known to induce the production of cytokines and chemokines in these cells, cytoplasmic IκB-α becomes degraded and NF-κB/Rel proteins translocate to nuclear fractions, resulting in an increased nuclear NF-κB DNA binding activity. Moreover, these various events are paralleled by the accumulation of IκB-α mRNA transcripts, which eventually leads to the protein synthesis-dependent reexpression of the IκB-α protein. To our knowledge, this represents the first indication that NF-κB activation may underlie the action of proinflammatory stimuli towards human neutrophil gene expression.
MATERIALS AND METHODS
Antibodies and reagents.Rabbit antisera to human c-Rel (no. 265 and no. 1136, raised against the C-terminal region and against an internal sequence downstream from the nuclear localization signal, respectively), p65/RelA (no. 1207 and no. 1226, against the N-terminal and C-terminal regions, respectively), p50/NFκB1 (no. 1141, against the N-terminal region, and no. 1157, against amino acids 339-357) and p52/NFκB2 (no. 1267, against the N-terminal region), were a generous gift from Dr N.R. Rice (NCI-Frederick Cancer Research and Development Center, Frederick, MD). The specificity of these antisera has already been extensively characterized.28 29 Recombinant human IκB-α30 was kindly provided by Dr R.T. Hay (Division of Cell and Molecular Biology, University of St Andrews, Fife, Scotland), and a rabbit polyclonal antibody (no. 9) raised against an N-terminal IκB-α peptide (ERPQEWAMEGPRDGL), was a kind gift from Dr S. Haskill (Department of Microbiology and Immunology, University of North Carolina, Chapel Hill, NC). A purified antibody to human IκB-α (sc-371) was also purchased from Santa Cruz Biotechnology Inc (Santa Cruz, CA). Rabbit antisera raised against leukotriene (LT)A4 hydrolase and five lipoxygenase-activating protein (FLAP) were generously provided by Dr Jilly Evans of Merck-Frosst Canada (Pointe-Claire, Québec, Canada). An oligonucleotide containing tandemly repeated NF-κB sites identical to those of the human immunodeficiency virus (HIV) promoter (5′-GATCA GGGACTTTCCGCTGGGGACTTTCC-3′; underlined is the NF-κB binding sequence) was kindly provided by Dr G. Trinchieri (Wistar Institute, Philadelphia, PA). An identical oligonucleotide containing mutated NF-κB sites (underlined are the altered nucleotides: 5′-AATACTTTCC) was synthesized by Dr Marcello Merola (Department of Biochemistry, University of Verona, Verona, Italy). Protein-G sepharose 4FF, Ficoll-Paque, T4 polynucleotide kinase, poly (dI-dC), and Sephadex G25 spin columns were purchased from Pharmacia (Uppsala, Sweden). Horseradish peroxidase-linked donkey antirabbit antibody, the protein biotinylation kit, the enhanced chemoluminescence (ECL) detection kit, and [γ-32P]-adenosine triphosphate (ATP) were purchased from Amersham (Little Chalfont, England). RPMI 1640 was from GIBCO-BRL (Gaithersburg, MD), and low-endotoxin fetal calf serum (FCS) (<6 pg/mL) from Hyclone (Logan, UT). Recombinant human (rh) TNF-α was purchased from Bachem Inc (Hannover, Germany), rhIL-1β from Hazleton Laboratories (Vienna, VA), and rh granulocyte colony-stimulating factor (G-CSF ) from R&D Systems Inc (Minneapolis, MN). Rh granulocyte macrophage (GM)-CSF and rhIL-8 were generous gifts from the Genetics Institute (Boston, MA), and Dr M. Ceska (Sandoz Research Institute Inc, Vienna, Austria), respectively; rhIFN-γ and rhIL-10 were kindly provided by Dr G. Garotta (Hoffmann-LaRoche, Basel, Switzerland) and Dr K. Moore (DNAX and Schering-Plough Corp, Palo Alto, CA), respectively. Polystyrene flasks and plates for cell culture were from Greiner (Nurtingen, Germany). Aprotinin, acetylated bovine serum albumin (BSA), cycloheximide, diisopropyl fluorophosphate (DFP), N-formyl-methionyl-leucyl-phenylalanine (fMLP), leupeptin, LPS, LTB4 , Nonidet P40 (NP40), pepstatin A, PAF, Percoll, phorbol 12-myristate 13-acetate (PMA), and phenylmethylsulfonyl fluoride (PMSF ) were all from the Sigma Chemical Co (St Louis, MO). Phosphoramidon and bestatin were from Boehringer-Mannheim (Mannheim, Germany). All other reagents were molecular biology grade, and all buffers and solutions were prepared using pyrogen-free clinical grade water.
Cell isolation and culture.Neutrophils were isolated from the peripheral blood of healthy donors under endotoxin-free conditions by a modification of the method of Boyum,31 as described earlier.2 Peripheral blood mononuclear cells were enriched in monocytes by centrifugation over Percoll cushions, as previously described.32 As determined by Wright staining and nonspecific esterase cytochemistry, the final neutrophil suspensions consistently contained fewer than 0.5% monocytes, and the monocyte-enriched suspensions contained 15% to 35% contaminating lymphocytes. Neutrophil and monocyte viability exceeded 98% after up to 3 hours in culture, as determined by trypan blue exclusion. Purified cell populations were resuspended in RPMI 1640 supplemented with 10% low-endotoxin FCS, at a final concentration of 5 × 106 cells/mL, and cultured in polystyrene flasks or in tubes at 37°C under a 5% CO2 atmosphere, with occasional agitation. Cell cultures were exposed to 1 μg/mL LPS, 100 U/mL TNF-α, or 10 nmol/L fMLP (or their diluent, RPMI 1640), for the indicated times. These concentrations of the agonists were chosen on the basis of their being optimal for cytokine and chemokine production in human neutrophils.2,33 34
Electrophoreses and immunoblots.After the desired incubation period with the stimuli, aliquots of the cell suspensions were transferred into precooled tubes containing equivalent volumes of ice-cold RPMI 1640 supplemented with DFP (2 mmol/L, final concentration) before centrifugation at 2,000g for 2 minutes at 4°C. The resulting cell pellets were washed and resuspended in ice-cold lysis buffer (10 mmol/L HEPES pH 7.90, 10 mmol/L NaCl, 1.5 mmol/L MgCl2 , 1 mmol/L EDTA, 0.5 mmol/L EGTA) containing 0.15% NP40 and an antiprotease cocktail (2 mmol/L DFP, 1 mmol/L PMSF, 1 mmol/L 4-(2-amino ethyl) benzene sulfonyl fluoride (AEBSF), and 10 μg/mL each of aprotinin, leupeptin and pepstatin A, final concentrations). Following a 10-minute incubation on ice, samples were briefly vortexed and centrifuged at 800g (10 minutes, 4°C). The supernatants (nonnuclear fractions) were collected, and the pellets (nuclear-containing fractions) were washed twice in lysis buffer containing the aforementioned antiprotease cocktail. Nonnuclear fractions were immediately dissolved twofold by the addition of an equal volume of sample buffer 2× (50 mmol/L TrisBase pH 6.80, 4% sodium dodecyl sulfate [wt/vol], 10% 2-mercaptoethanol [vol/vol], 20% glycerol [vol/vol]), heated for 5 minutes at 95°C, and used immediately or stored at −20°C. Washed nuclear fractions were resuspended in 100 μL of DNAse buffer (20 mmol/L HEPES pH 7.50, 1 mmol/L CaCl2 , 5 mmol/L MgCl2 , 1 mmol/L EDTA) containing 10 units of DNAse I and the aforementioned antiprotease cocktail, and incubated for 30 minutes at 37°C. Reactions were stopped by the addition of an equal volume of sample buffer 2×, and samples were heated for 5 minutes at 95°C and used immediately or stored at −20°C.
Samples were electrophoresed on 15% denaturing gels (or 18% gels in the case of IκB-α or FLAP), prepared according to the method of Thomas and Kornberg.35 Proteins were then transferred onto a nitrocellulose blotting membrane at 20 V constant for 60 minutes in a Transblot semidry transfer cell (BioRad, Hercules, CA). Transfer efficiency was visualized by reversible Ponceau Red staining. The membranes were first soaked for 60 minutes at 37°C in TBS (25 mmol/L Tris-HCl pH 7.60, 0.2 mol/L NaCl, 0.15% Tween 20) containing 2% gelatin (wt/vol) or 1.5% low-fat skimmed milk (depending on the primary antibody to be used), and subsequently exposed (60 minutes, 37°C) to a 1:1,500 dilution of either anti-p65 (no. 1207 or no. 1226), anti-p50, or anti-p52 antisera, a 1:2,000 dilution of anti-IκB–α peptide antibody (no. 9), a 1:300 dilution of commercial anti-IκB–α antibody (sc-371), or to a 1:20,000 dilution of anti c-Rel (no. 265) antiserum. Antisera to FLAP and to LTA4 hydrolase were used at 1:4,000 and 1:5,000 dilutions, respectively. The membranes were then washed three times with 150 mL TBS, and incubated in TBS with a horseradish peroxidase-linked donkey antirabbit antibody, added to a final dilution of 1:10,000, for 45 minutes at 37°C. After three washes, the signal was revealed with the ECL reagent, according to the manufacturer's instructions. As a positive migration control for NF-κB/Rel proteins, a track of whole-cell samples from unstimulated Jurkat T cells was usually included on the gels.
Immunoprecipitations.Nonnuclear fractions were prepared as described above, deposited onto Microcon-100 microcolumns (Amicon Inc, Beverly, MA), which have a molecular weight cut-off of approximately 100 kD, and centrifuged (3,000g, 30 minutes, 4°C). IκB-α was consistently undetectable in the eluates, as determined by immunoblot (data not shown). The retentates were flushed out of the microcolumns according to the manufacturer's instructions, using 100 μL of immunoprecipitation buffer (10 mmol/L Tris-HCl pH 7.40, 1% NP40, 150 mmol/L NaCl, 1 mmol/L EDTA) supplemented with the antiprotease cocktail. Sample volume was then brought to 475 μL with immunoprecipitation buffer, and immunoprecipitations were performed as follows. A total of 20 μL of a 1:2 (vol:vol) dilution of protein-G sepharose 4FF was mixed with preimmune rabbit serum (5 μL) for 30 minutes at room temperature. Preclearing of the samples was achieved by incubating (120 minutes at 4°C, under agitation) with the protein-G sepharose mixture. Samples were then centrifuged (12,000g, 2 minutes), and the protein-G pellets discarded. During this time, 5 μL of a 1:5 dilution of either anti-IκB-α antibody (no. 9), or antisera raised against individual Rel family proteins, was mixed with 20 μL of the protein-G sepharose mixture for 30 minutes at room temperature, before a 120-minute incubation with the supernatants (4°C, with agitation). Samples were then centrifuged (10 minutes, 12,000g ), and the resulting immunoprecipitates were washed three times with rinsing buffer (25 mmol/L Tris-HCl pH 8.0, 150 mmol/L NaCl, 0.1% NP40), before being dissolved in sample buffer and heated for 5 minutes at 95°C. Following electrophoresis and transfer onto nitrocellulose membranes, immunoblots were performed as described above.
Electrophoretic mobility shift assays (EMSA).Neutrophils were cultured in the presence or absence of various stimuli for the indicated times; incubations were stopped as described above, and the cells were pelleted and resuspended in ice-cold relaxation buffer (10 mmol/L PIPES pH 7.30, 30 mmol/L KCl, 3 mmol/L NaCl, 3.5 mmol/L MgCl2 , 1.25 mmol/L EGTA, 0.5 mmol/L DTT) containing the antiprotease cocktail. Cells were disrupted by nitrogen cavitation, using a modification of a previously published procedure.36 Briefly, neutrophils (108 cells/mL) were pressurized under a N2 atmosphere (350 psi, 20 minutes at 4°C) with constant stirring in a nitrogen bomb (Parr Instrument Co, Moline, IL). Cavitates were spun at 1,000g (10 minutes, 4°C), to pellet unbroken cells and intact nuclei, and the supernatants were recentrifuged (1,000g, 10 minutes, 4°C) to remove remaining nuclei and unbroken cells. Both pellets were combined and resuspended in 200 μL relaxation buffer containing the antiprotease cocktail; the resulting suspension was laid onto 2 × 400 μL of a Percoll step gradient (1.050 and 1.120 g/mL), and centrifuged at 2,200g (10 minutes, 4°C). The top of the gradient (containing the unbroken cells) was discarded, and the nuclei-rich fraction at the interface of the two Percoll layers was carefully collected, diluted in relaxation buffer containing the antiprotease cocktail, and repelleted (1,500g, 10 minutes, 4°C), to rid the nuclei of remaining Percoll. The 1,000-g (postnuclear) supernatants were centrifuged at 13,000g (15 minutes, 4°C) to pellet neutrophil granules. The resulting 13,000-g supernatants are referred to as “cytoplasmic” fractions, even though they do contain plasma membranes in addition to cytosol.
Nuclear and cytoplasmic extracts were prepared by a modified Dignam procedure.37 Washed nuclear pellets were gently resuspended in ice-cold relaxation buffer containing 10% (vol/vol) glycerol and the antiprotease cocktail; NaCl was then added to yield a final concentration of 380 mmol/L. Following a 20-minute incubation on ice (with occasional mixing), samples thus treated were spun at 13,000g (15 minutes, 4°C), and the resulting supernatants (the nuclear extracts) were aliquoted and immediately stored at −70°C. Similarly, cytoplasmic fractions were supplemented with glycerol (10% final concentration) and with concentrated NaCl (to yield a final concentration of 150 mmol/L), and incubated on ice for 20 minutes before centrifugation at 13,000g (15 minutes, 4°C). The resulting supernatants, referred to as cytoplasmic extracts, were aliquoted and stored at −70°C before use. In some experiments, whole-cell extracts were prepared by processing neutrophil cavitates in the same manner as described above for cytoplasmic extracts. Small aliquots of the various extracts were routinely processed for protein content determination.
EMSA was performed essentially as described earlier,38 with the following modifications. Binding reactions were performed in 20 μL binding buffer (20 mmol/L Tris-HCl pH 7.50, 50 mmol/L KCl, 1 mmol/L EDTA, 1 mmol/L DTT, 0.1% (vol/vol) NP40, 6% glycerol) supplemented with 20 μg acetylated BSA and 1 μg poly (dI-dC). Extracts (amounts used are indicated in the figure legends) were then added to the binding mixture and allowed to equilibrate for 15 minutes at room temperature. In some instances, binding mixtures containing cytoplasmic or whole-cell extracts were sequentially treated (10 minutes, room temperature) with 0.7% sodium deoxycholate (DOC) (vol/vol) and 1% NP-40 (vol/vol), to dissociate NF-κB complexes from endogenous inhibitors.20 For competition or supershift experiments, binding reactions were performed in the presence of cold competitors or of specific antisera, for 20 minutes at 4°C. Finally, 40,000 cpm of an oligonucleotide containing the consensus NF-κB sequence, 32P-end–labeled using T4 polynucleotide kinase, was added to the binding mixtures, which were further incubated for 15 minutes at room temperature. The resulting samples were electrophoresed on 6% native gels at 4°C in 0.5× TBE.
Isolation of mRNA and Northern blots.Total RNA was extracted and analyzed by Northern blotting as described previously.2 For this purpose, an IκB-α cDNA fragment was [α-32P]-dCTP–labeled with a Ready-to-Go DNA labeling kit (Pharmacia, Uppsala, Sweden), before hybridization on nylon filters and autoradiography. This IκB-α cDNA fragment was prepared by reverse transcriptase-polymerase chain reaction (RT-PCR) amplification of mRNA purified from LPS-stimulated monocytes, using oligonucleotide primers that are specific for IκB-α (kindly provided by Dr S. Haskill). Contaminating monocyte mRNA was undetectable in our neutrophil preparations, as the filters did not hybridize with an IL-6 cDNA probe.2 39
RESULTS
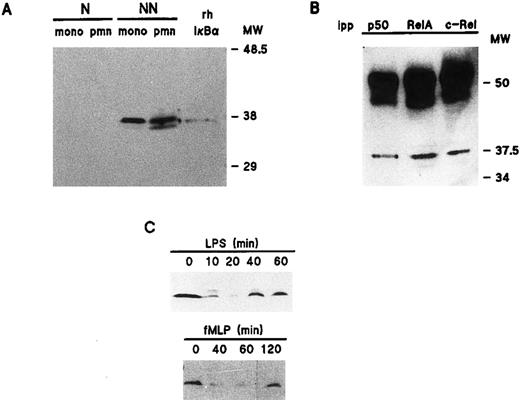
Relative abundance and distribution of NF-κB/Rel proteins in resting human neutrophils and monocytes.To determine which Rel family proteins are present in human neutrophils, as well as their distribution in resting cells, subcellular fractions obtained by NP40 lysis were electrophoresed and processed for immunoblotting using specific antisera to individual NF-κB/Rel proteins. For comparative purposes, corresponding fractions prepared using monocyte-enriched suspensions from the same donors were analyzed in parallel. Figure 1A shows that in addition to abundant quantities of c-Rel, whose presence in neutrophils has already been reported,40 neutrophils were also found to contain p50/NFκB1 and its precursor, p105, as well as p65/RelA. In contrast, neither p52/NFκB2 nor its precursor, p100, were detectable in neutrophils, even when 4 million cell-equivalents were loaded on the gels (data not shown). By comparison, monocyte-enriched suspensions contained substantial amounts of p52 and p100 (Fig 1A). Noteworthy is that in neutrophils, immunoblot detection of p65 (but not that of c-Rel or p50) using both the C-terminal and N-terminal antisera required the presence of DFP in the various buffers used during sample preparation (data not shown), in agreement with recent observations made in certain U937 cell subclones containing elevated levels of intracellular proteases that are typically found in granulocytes.41
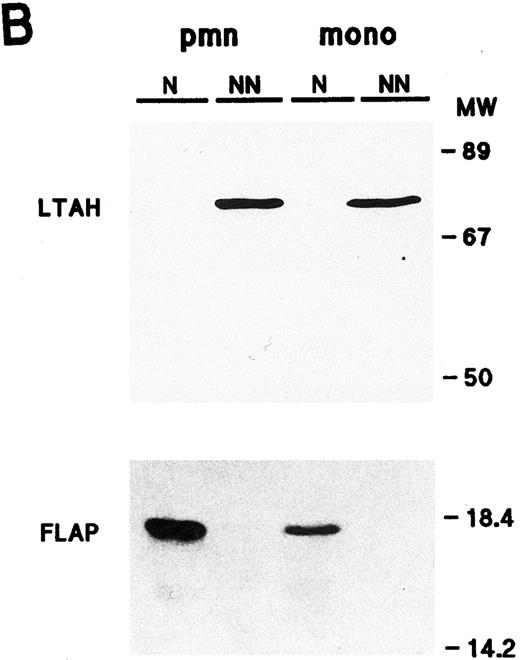
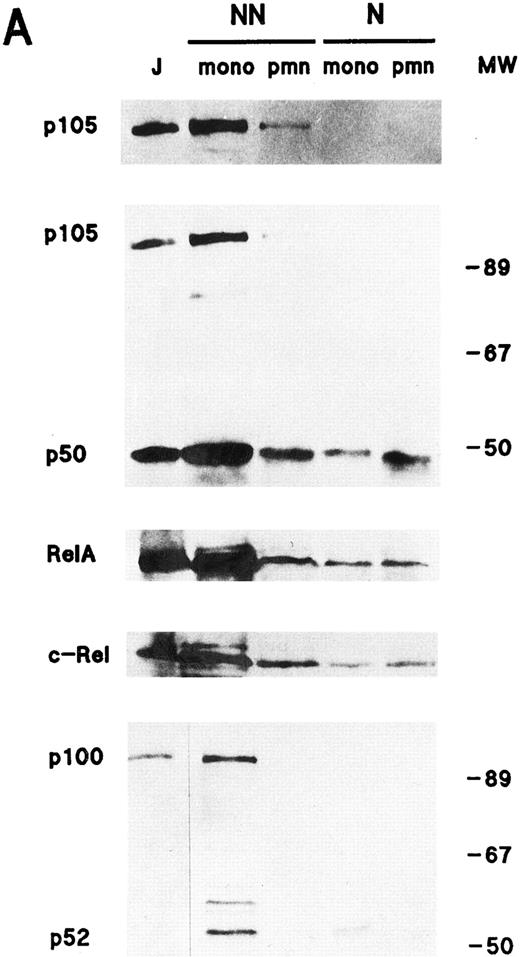
Cellular distribution of NF-κB/Rel proteins in peripheral blood neutrophils and monocytes. (A) Unstimulated neutrophils or monocyte-enriched mononuclear cell suspensions from the same donor were submitted to NP40 lysis, and the resulting nuclear-containing and nonnuclear fractions were processed for electrophoresis on 15% Kornberg gels (using 106 cell-equivalents per well, unless otherwise stated) and immunoblotting, as described in Materials and Methods. For p105 and p50, the immunoblot was performed using the no. 1157 anti-p50 antiserum; to allow a better visualization of the p105 band in neutrophil nonnuclear fractions, a longer exposure of the same film is shown in the top panel. For p65/RelA, the immunoblot was performed using an antiserum to the N-terminal region (no. 1207). For c-Rel, 6 × 105 cell-equivalents per well were loaded on the gel and immunoblots were performed using the no. 265 antiserum. As a positive migration control for NF-κB/Rel proteins, a whole-cell sample from unstimulated Jurkat T cells was loaded on the outermost left track (“J”). This experiment is representative of at least three. (B) Immunoblots were performed on the same samples as the ones depicted in (A). Upper panel, 106 cell-equivalents were loaded on a 15% Kornberg gel and the membrane was immunoblotted using an anti-LTA4 hydrolase antiserum (“LTAH”). Lower panel, 5 × 105 cell-equivalents were loaded on an 18% Kornberg gel and the membrane was immunoblotted using an anti-FLAP antiserum. This experiment is representative of at least three. N, nuclear fractions; NN, nonnuclear fractions; pmn, polymorphonuclear neutrophils; mono, autologous monocyte-enriched suspensions; MW, molecular weight markers (in kD).
Cellular distribution of NF-κB/Rel proteins in peripheral blood neutrophils and monocytes. (A) Unstimulated neutrophils or monocyte-enriched mononuclear cell suspensions from the same donor were submitted to NP40 lysis, and the resulting nuclear-containing and nonnuclear fractions were processed for electrophoresis on 15% Kornberg gels (using 106 cell-equivalents per well, unless otherwise stated) and immunoblotting, as described in Materials and Methods. For p105 and p50, the immunoblot was performed using the no. 1157 anti-p50 antiserum; to allow a better visualization of the p105 band in neutrophil nonnuclear fractions, a longer exposure of the same film is shown in the top panel. For p65/RelA, the immunoblot was performed using an antiserum to the N-terminal region (no. 1207). For c-Rel, 6 × 105 cell-equivalents per well were loaded on the gel and immunoblots were performed using the no. 265 antiserum. As a positive migration control for NF-κB/Rel proteins, a whole-cell sample from unstimulated Jurkat T cells was loaded on the outermost left track (“J”). This experiment is representative of at least three. (B) Immunoblots were performed on the same samples as the ones depicted in (A). Upper panel, 106 cell-equivalents were loaded on a 15% Kornberg gel and the membrane was immunoblotted using an anti-LTA4 hydrolase antiserum (“LTAH”). Lower panel, 5 × 105 cell-equivalents were loaded on an 18% Kornberg gel and the membrane was immunoblotted using an anti-FLAP antiserum. This experiment is representative of at least three. N, nuclear fractions; NN, nonnuclear fractions; pmn, polymorphonuclear neutrophils; mono, autologous monocyte-enriched suspensions; MW, molecular weight markers (in kD).
In both neutrophils and monocytes, a greater proportion of the Rel family proteins was recovered in the non-nuclear fractions (Fig 1A). A similar cellular distribution was observed in resting neutrophils disrupted by nitrogen cavitation instead of detergent lysis, with the exception that nuclear levels of Rel proteins were less abundant using the cavitation procedure (data not shown). The presence of detectable quantities of p50, p65, and c-Rel (but not p100 or p105) in the nuclear fractions of neutrophils and monocytes, however, prompted us to ascertain the identity and purity of our subcellular fractions. For this purpose, we examined their respective FLAP and LTA4 hydrolase content (Fig 1B). FLAP is an 18-kD protein that is known to be exclusively located in the nuclear envelope of human neutrophils and monocytes,42 while LTA4 hydrolase, a protein of approximately 70 kD, is strictly cytosolic, regardless of whether cells are fractionated by sonication43 or nitrogen cavitation (unpublished data, October 1995). That FLAP was only detected in nuclear fractions, and LTA4 hydrolase in nonnuclear fractions, demonstrates that these subcellular fractions are reasonably exempt from cross-contamination. Interestingly, neutrophils were found to contain more FLAP than autologous monocytes, in agreement with previous observations.44 Another noticeable difference between neutrophils and monocytes is that the latter contain more cytoplasmic NF-κB/Rel proteins than the former. This was particularly evident in the case of p52/p100, as already mentioned. Conversely, neutrophils were found to contain more nuclear p50 and c-Rel than monocytes. Collectively, these differences indicate that results obtained using our neutrophil preparations do not reflect a contribution of contaminating monocytes, in agreement with the fact that monocytes account for less than 0.5% of the cells (as determined by esterase cytochemistry).
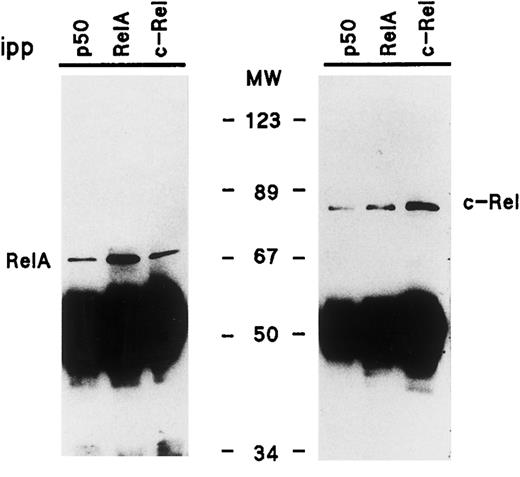
We also investigated the physical association of NF-κB/Rel proteins with one another in nonnuclear fractions of resting neutrophils. For this purpose, the fractions were immunoprecipitated using antisera to individual NF-κB/Rel proteins, and the resulting immunoprecipitates were immunoblotted for c-Rel or p65/RelA. Figure 2 shows that in addition to being efficiently precipitated by a combination of anti-p65 antisera (no. 1207 and no. 1226), the p65/RelA protein could also be coimmunoprecipitated using antisera raised against either c-Rel or p50. Similarly, c-Rel was not only immunoprecipitated using an antiserum to itself, but also by either anti-p50 or anti-p65 antisera (Fig 2). In addition, immunoblot analysis of the same immunoprecipitates using an anti-p50 antiserum (no. 1157) showed that while the antiserum efficiently brought down the p105 protein, the p105 signal was consistently below detection in anti-cRel and anti-RelA immunoprecipitates (data not shown); under these conditions, the immunoglobulin heavy chain strongly interfered with the detection of p50. This indicates that very little, if any, cytoplasmic RelA or c-Rel is complexed to p105. Therefore, the immunoprecipitable RelA or c-Rel brought down using the anti-p50 antiserum must principally reflect heterodimers containing p50 (as opposed to p105), consistent with the fact that neutrophils contain considerably more p50 than p105 (Fig 1A). Similar results were obtained using cytosolic fractions from neutrophils disrupted by nitrogen cavitation instead of NP40 lysis (data not shown). Taken together, these experiments clearly demonstrate the presence of p50/p65, p50/c-Rel, and p65/c-Rel heterodimers in resting neutrophil cytoplasm.
Characterization of the NF-κB/Rel protein heterodimers present in nonnuclear fractions of resting human neutrophils. Following NP40 lysis of neutrophils, nonnuclear fractions were immunoprecipitated using antisera to individual NF-κB/Rel proteins, and immunoprecipitates (2.5 × 106 cell-equivalents) were analyzed by immunoblot using an antiserum to the N-terminal region of p65/RelA (no. 1207, left panel), or an antiserum to c-Rel (no. 265, right panel). This experiment is representative of at least three. MW, molecular weight markers (in kD); ipp, antiserum used for immunoprecipitation.
Characterization of the NF-κB/Rel protein heterodimers present in nonnuclear fractions of resting human neutrophils. Following NP40 lysis of neutrophils, nonnuclear fractions were immunoprecipitated using antisera to individual NF-κB/Rel proteins, and immunoprecipitates (2.5 × 106 cell-equivalents) were analyzed by immunoblot using an antiserum to the N-terminal region of p65/RelA (no. 1207, left panel), or an antiserum to c-Rel (no. 265, right panel). This experiment is representative of at least three. MW, molecular weight markers (in kD); ipp, antiserum used for immunoprecipitation.
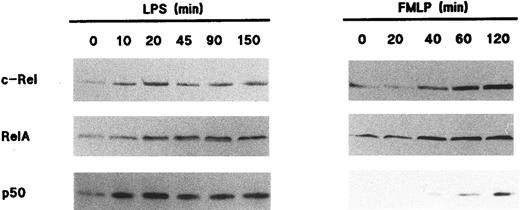
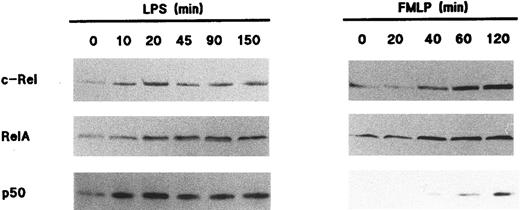
Redistribution of Rel family proteins following neutrophil activation.To investigate the fate of NF-κB/Rel proteins following cell activation, neutrophils were stimulated with various agonists, and the resulting nuclear fractions were analyzed by immunoblot. Figure 3 shows that in response to 1 μg/mL LPS, c-Rel, p65/RelA, and p50 translocated to the nuclear fractions; this was paralleled by a decrease in the amount of these proteins in the corresponding nonnuclear fractions (data not shown). The effect of LPS towards the translocation of Rel family proteins was rapid, as it was detectable within 10 minutes and was sustained for up to 2.5 hours. In this respect, the elevated nuclear levels of all three Rel proteins were indistinguishable between NP40-disrupted and cavitated neutrophils following LPS stimulation (data not shown). Stimulation of neutrophils with 100 U/mL TNF-α yielded qualitatively identical results with respect to both the extent and time course of NF-κB/Rel protein translocation (data not shown). Interestingly, a redistribution of NF-κB/Rel proteins was also observed in fMLP-stimulated neutrophils, although it required approximately 45 minutes to be evident (Fig 3). Whereas c-Rel and p65/RelA were efficiently mobilized to the nucleus in response to fMLP, it must be stressed that under the same conditions, only a minor fraction of cytoplasmic p50 translocated to nuclear fractions. In five independent experiments, this modest translocation of the p50 protein was clearly observed in three donors (as depicted in Fig 3), whereas it was less evident in the other two. Among several other neutrophil agonists that were tested for their ability to promote the nuclear mobilization of Rel family proteins, PMA stood out as a relatively potent inducer, behaving in a similar manner to fMLP in terms of the kinetics of this response (data not shown). Table 1 summarizes the effect of various neutrophil agonists towards the subcellular distribution of Rel family proteins.
Nuclear mobilization of Rel family proteins following stimulation of human neutrophils. Cells (5 × 106/mL) were cultured at 37°C in the presence of 1 μg/mL LPS (left panel) or 10 nmol/L fMLP (right panel); aliquots were taken at the indicated times (in minutes), and submitted to NP40 lysis. The resulting nuclear fractions were then analyzed by immunoblot using antisera to p50 (no. 1141), c-Rel, or p65/RelA (no. 1207); 106 cell-equivalents were loaded on the gels (except when probed for c-Rel, 7 × 105 cell-equivalents). The experiments depicted in this figure are representative of at least five.
Nuclear mobilization of Rel family proteins following stimulation of human neutrophils. Cells (5 × 106/mL) were cultured at 37°C in the presence of 1 μg/mL LPS (left panel) or 10 nmol/L fMLP (right panel); aliquots were taken at the indicated times (in minutes), and submitted to NP40 lysis. The resulting nuclear fractions were then analyzed by immunoblot using antisera to p50 (no. 1141), c-Rel, or p65/RelA (no. 1207); 106 cell-equivalents were loaded on the gels (except when probed for c-Rel, 7 × 105 cell-equivalents). The experiments depicted in this figure are representative of at least five.
In a separate series of experiments, it was determined that a 30-minute preincubation of neutrophils with 20 μg/mL cycloheximide had no inhibitory effect towards the nuclear accumulation of Rel family proteins observed in response to LPS, TNF-α, or fMLP (data not shown). Thus, the inducible redistribution of NF-κB/Rel proteins observed in human neutrophils is a process that does not require de novo protein synthesis. In the same experiments, cycloheximide pretreatment also failed to affect the total cellular pool of immunoreactive Rel family proteins, regardless of whether the cells were subsequently stimulated (for up to 2.5 hours) with either LPS, TNF-α, or fMLP. Therefore, this indicates that Rel family proteins do not have a rapid turnover in human neutrophils.
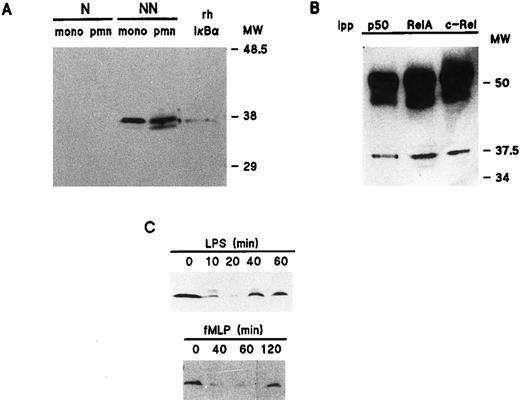
Distribution and inducible degradation of the IκB-α protein in human neutrophils.We next examined the relative abundance of IκB-α in unstimulated neutrophils and autologous monocyte-enriched suspensions. Figure 4A shows that in both cell types, a major immunoreactive band was exclusively detected in the nonnuclear fractions, which comigrated with authentic rhIκB-α. The identity of this band was ascertained in control experiments by preincubating our anti–IκB-α antibody with the recombinant protein, a treatment which prevented its subsequent detection by immunoblot (data not shown). Conversely, a nonspecific band that was sometimes detected right below the authentic IκB-α band could not be displaced by this treatment. We next investigated the physical association of IκB-α with Rel family proteins in resting neutrophils. To this end, nonnuclear fractions from resting neutrophils were immunoprecipitated with antisera to individual NF-κB/Rel proteins and immunoblotted using an anti–IκB-α antibody. As shown in Fig 4B, this allowed the detection of IκB-α in all immunoprecipitates. Thus, it appears that in resting neutrophils, IκB-α is complexed with protein dimers consisting of at least one of the three Rel family proteins that are present in these cells. Further support for this conclusion is that both RelA and c-Rel were detected by immunoblot analysis of neutrophil nonnuclear fractions immunoprecipitated using an anti–IκB-α antibody (data not shown).
Cellular distribution and inducible degradation and resynthesis of IκB-α in human neutrophils. (A) Unstimulated neutrophils or monocyte-enriched mononuclear cell suspensions from the same donor were submitted to NP40 lysis, and the resulting nuclear-containing (“N”) and nonnuclear (“NN”) fractions were processed for electrophoresis (2 × 106 cell-equivalents per well) and immunoblotting using an anti-IκB–α antibody (no. 9). For comparative purposes, the immunoblot shown in this panel was performed using samples from the same experiment as the one depicted in Fig 1. This experiment is representative of three. (B) Nonnuclear fractions from resting neutrophils were immunoprecipitated with antisera raised against individual NF-κB/Rel proteins, and the resulting immunoprecipitates (2.5 × 106 cell-equivalents) were processed for immunoblot analysis using an anti-IκB–α antibody (sc-371). For comparative purposes, the immunoblot shown in this panel was performed using samples from the same experiment as the one depicted in Fig 2. This experiment is representative of two. (C) Neutrophils (5 × 106/mL) were cultured at 37°C in the presence of 1 μg/mL LPS or 10 nmol/L fMLP. Aliquots were taken at the indicated times, submitted to nonionic detergent lysis, and the resulting nonnuclear fractions were processed for immunoblot analysis using an anti-IκB–α antibody (sc 371). Each of the experiments depicted in this panel is representative of at least four. pmn, polymorphonuclear neutrophils; mono, autologous monocyte-enriched suspensions; MW, molecular weight markers (in kD); ipp, antiserum used for immunoprecipitation.
Cellular distribution and inducible degradation and resynthesis of IκB-α in human neutrophils. (A) Unstimulated neutrophils or monocyte-enriched mononuclear cell suspensions from the same donor were submitted to NP40 lysis, and the resulting nuclear-containing (“N”) and nonnuclear (“NN”) fractions were processed for electrophoresis (2 × 106 cell-equivalents per well) and immunoblotting using an anti-IκB–α antibody (no. 9). For comparative purposes, the immunoblot shown in this panel was performed using samples from the same experiment as the one depicted in Fig 1. This experiment is representative of three. (B) Nonnuclear fractions from resting neutrophils were immunoprecipitated with antisera raised against individual NF-κB/Rel proteins, and the resulting immunoprecipitates (2.5 × 106 cell-equivalents) were processed for immunoblot analysis using an anti-IκB–α antibody (sc-371). For comparative purposes, the immunoblot shown in this panel was performed using samples from the same experiment as the one depicted in Fig 2. This experiment is representative of two. (C) Neutrophils (5 × 106/mL) were cultured at 37°C in the presence of 1 μg/mL LPS or 10 nmol/L fMLP. Aliquots were taken at the indicated times, submitted to nonionic detergent lysis, and the resulting nonnuclear fractions were processed for immunoblot analysis using an anti-IκB–α antibody (sc 371). Each of the experiments depicted in this panel is representative of at least four. pmn, polymorphonuclear neutrophils; mono, autologous monocyte-enriched suspensions; MW, molecular weight markers (in kD); ipp, antiserum used for immunoprecipitation.
To determine the fate of IκB-α following cell activation, neutrophils were stimulated under the same conditions that were found to promote the nuclear translocation of Rel family proteins. Figure 4C shows that in cells stimulated with 1 μg/mL LPS, cytoplasmic IκB-α levels had substantially decreased by 10 minutes, reaching a minimum after 20 to 30 minutes (depending on the donor). This gradual loss of immunoreactive IκB-α protein was accompanied by the detection of a slower-migrating band, which presumably represents a hyperphosphorylated IκB-α species, based on similar observations made in other cell types.21 Interestingly, we repeatedly observed that a weak residual amount of IκB-α protein was still detectable following cell stimulation; we do not know at present whether this reflects an incomplete degradation of IκB-α or the rapid onset of de novo protein synthesis. Nevertheless, evidence in favor of the latter mechanism is that the loss of IκB-α was invariably followed by an increase in the level of the protein, which was usually detectable after approximately 40 minutes of stimulation, and the cellular pool of IκB-α was almost replenished by 60 minutes (Fig 4C). Further evidence is that exposure of neutrophils to 20 μg/mL cycloheximide for 30 minutes before LPS stimulation prevented this reappearance of IκB-α (data not shown), although a weak residual amount of IκB-α protein was often detectable following cycloheximide treatment. Taken together, these data indicate that IκB-α reexpression is mostly dependent on de novo protein synthesis. In neutrophils stimulated with 100 U/mL TNF-α, IκB-α was degraded and reexpressed with similar kinetics as in LPS-treated cells (data not shown). By comparison, the loss of immunoreactive IκB-α protein was only evident after 30 to 60 minutes (depending on the donor) in neutrophils stimulated with 10 nmol/L FMLP (Fig 4C). In five independent experiments, cytoplasmic levels of IκB-α had returned to near basal levels after 120 minutes in three donors (as depicted in Fig 4C), whereas the reappearance of the protein was only observed after 180 minutes in the other two donors.
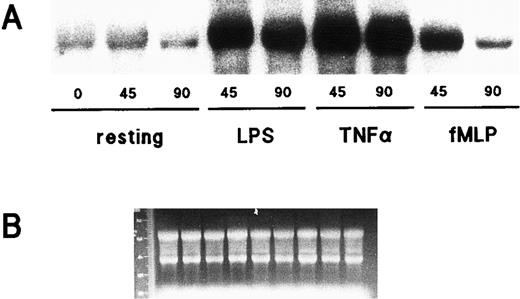
Accumulation of IκB-α mRNA following neutrophil activation.That IκB-α protein is synthesized de novo following neutrophil stimulation with LPS, TNF-α, or fMLP, prompted us to investigate whether this response might be paralleled by changes in IκB-α mRNA steady-state levels. Neutrophils were, therefore, stimulated for varying lengths of time with either LPS, TNF-α, or fMLP; total RNA was then extracted and processed for Northern blot analysis. Figure 5 shows that a low quantity of IκB-α transcripts was consistently detected in resting neutrophils; following cell exposure to either LPS or TNF-α, a dramatic and sustained accumulation of IκB-α mRNA was observed. In neutrophils thus stimulated, the steady-state level of IκB-α mRNA remained elevated even after 3 hours of stimulation, relative to unstimulated controls (data not shown). By comparison, fMLP induced a less marked and more transient accumulation of IκB-α transcripts (Fig 5). This effect of fMLP could be detected as early as 30 minutes and had become completely undetectable by 120 minutes (data not shown). In a separate series of experiments, neutrophils were pretreated with either 5 μg/mL actinomycin D or 20 μg/mL cycloheximide (or their respective diluents) for 30 minutes at 37°C before stimulation with LPS or fMLP for 45 minutes at 37°C. Actinomycin D was found to block the accumulation of IκB-α transcripts, whereas cycloheximide superinduced it (data not shown). Both compounds exerted a similar effect on the steady-state level of IκB-α mRNA in unstimulated cells (data not shown). Thus, it appears that while the inducible accumulation of IκB-α mRNA involves transcriptional events, it does not require the synthesis of proteic factors.
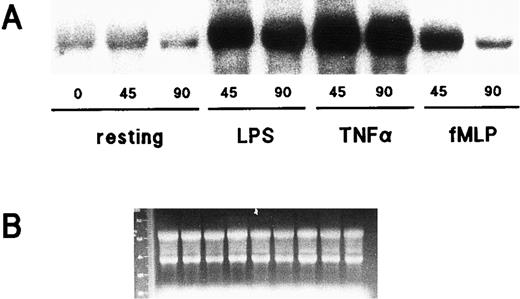
Effect of LPS, TNF-α, and fMLP on IκB-α gene expression in human neutrophils. (A) Cells (5 × 106/mL) were cultured at 37°C in the presence or absence of 1 μg/mL LPS, 5 ng/mL TNF-α, or 10 nmol/L fMLP for the indicated times (in minutes). Total RNA was then extracted and processed for Northern blot analysis using an IκB-α cDNA probe. This experiment is representative of three. (B) Ethidium bromide staining of the gel corresponding to the autoradiograms depicted in (A).
Effect of LPS, TNF-α, and fMLP on IκB-α gene expression in human neutrophils. (A) Cells (5 × 106/mL) were cultured at 37°C in the presence or absence of 1 μg/mL LPS, 5 ng/mL TNF-α, or 10 nmol/L fMLP for the indicated times (in minutes). Total RNA was then extracted and processed for Northern blot analysis using an IκB-α cDNA probe. This experiment is representative of three. (B) Ethidium bromide staining of the gel corresponding to the autoradiograms depicted in (A).
Induction of NF-κB DNA binding activity following neutrophil activation.That neutrophils can be induced to mobilize NF-κB/Rel proteins to the nucleus strongly suggested that this response, as well as the related cellular events that occur under the same conditions, might be paralleled by NF-κB activation. Figure 6A shows that whole-cell extracts from resting neutrophils contained a latent NF-κB DNA binding activity (complex A) that became detectable following DOC treatment, as determined by EMSA. Subcellular fractionation of neutrophils further showed that this DOC-releasable NF-κB DNA binding activity was restricted to cytoplasmic extracts (data not shown), in agreement with the fact that IκB-α is exclusively detected in neutrophil cytoplasmic fractions. In addition, a weak constitutive NF-κB DNA binding activity was usually detectable in whole-cell extracts from resting cells, although this generally required longer exposures to autoradiographic film; this constitutive complex was assigned to the corresponding nuclear fractions (see below). By comparison, a DNA binding activity of similar electrophoretic mobility (complex A) was readily detected in whole-cell extracts from LPS-treated neutrophils (Fig 6A). DOC treatment of these extracts only resulted in a moderate increase in NF-κB DNA binding activity. Collectively, these observations suggested that a predominant part of the cellular NF-κB pool had translocated to the nucleus in response to LPS stimulation. Accordingly, the inducible NF-κB DNA binding activity present in whole-cell extracts of LPS-treated neutrophils was detected in nuclear extracts prepared from the same cells, but not in the corresponding cytoplasmic extracts (Fig 6B). Conversely, two faster-migrating DNA binding activities (complexes B and C), which were also present in neutrophil whole-cell extracts and which strongly bound to our NF-κB probe, were almost entirely recovered in the corresponding cytoplasmic extracts (Fig 6B), and were not significantly affected by LPS challenge. Similar results were obtained in TNF-α–stimulated neutrophils, or when the various extracts were analyzed using an oligonucleotide probe containing the κB motif present in the Ig κ promoter (data not shown).
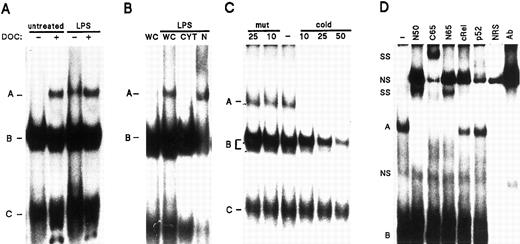
Detection and characterization of NF-κB DNA binding activities in human neutrophils. (A) Neutrophils (5 × 106/mL) were cultured in the presence or absence of 1 μg/mL LPS, for 15 minutes at 37°C. Whole-cell extracts were prepared as described in Materials and Methods, and incubated in the presence or absence of DOC in the binding mixtures, before EMSA analysis using an oligonucleotide probe containing tandem repeats of the consensus NF-κB motif. The various DNA-binding complexes are indicated by the letters, A, B, and C. The amount of extract used in the binding reactions corresponded to 620,000 cell equivalents (approximately 20 μg of protein). (B) Neutrophils were cultured in the absence (first lane) or presence of LPS as indicated in (A), and whole-cell extracts (“WC”), as well as the corresponding cytoplasmic (“CYT”) and nuclear (“N”) extracts, were analyzed in EMSA. The amount of extract used corresponded to 550,000 cell equivalents (approximately 19 μg, 18 μg, and 2.2 μg for whole-cell, cytoplasmic, and nuclear extracts, respectively). (C) Whole-cell extracts from neutrophils cultured in the presence of 100 U/mL TNF-α (20 minutes at 37°C) were prepared and analyzed in EMSA. Before addition of the labeled NF-κB oligonucleotide probe to the binding mixture, extracts were incubated with a 10-, 25- or 50-fold molar excess of unlabeled NF-κB oligonucleotide (“cold”), or with an excess of unlabeled oligonucleotide containing mutated NF-κB binding sites (“mut”). The amount of extract used corresponded to 510,000 cell equivalents (approximately 16 μg of protein). (D) Neutrophils were cultured in the presence of LPS as described in (A), and nuclear extracts were analyzed in EMSA. Before addition of the labeled NF-κB oligonucleotide probe, extracts were incubated with antisera raised against p50 (no. 1141), p52 (no. 1267), c-Rel (no. 1136), the C-terminal region of p65/RelA (“C65”, no. 1226), the N-terminal region of p65 (“N65”, no. 1207), or without antisera (“-”). As a control, binding mixtures that only contained one of the antisera (in this case the anti-c-Rel, “Ab”), or normal rabbit serum (“NRS”), were routinely included on the gels. The amount of extract used corresponded to 960,000 cell equivalents, representing about 4.8 μg of protein. Supershifted complexes (“SS”) are indicated, as well as nonspecific bands (“NS”) present either in normal rabbit serum or in the control antiserum. Each of the experiments depicted in this figure is representative of at least four.
Detection and characterization of NF-κB DNA binding activities in human neutrophils. (A) Neutrophils (5 × 106/mL) were cultured in the presence or absence of 1 μg/mL LPS, for 15 minutes at 37°C. Whole-cell extracts were prepared as described in Materials and Methods, and incubated in the presence or absence of DOC in the binding mixtures, before EMSA analysis using an oligonucleotide probe containing tandem repeats of the consensus NF-κB motif. The various DNA-binding complexes are indicated by the letters, A, B, and C. The amount of extract used in the binding reactions corresponded to 620,000 cell equivalents (approximately 20 μg of protein). (B) Neutrophils were cultured in the absence (first lane) or presence of LPS as indicated in (A), and whole-cell extracts (“WC”), as well as the corresponding cytoplasmic (“CYT”) and nuclear (“N”) extracts, were analyzed in EMSA. The amount of extract used corresponded to 550,000 cell equivalents (approximately 19 μg, 18 μg, and 2.2 μg for whole-cell, cytoplasmic, and nuclear extracts, respectively). (C) Whole-cell extracts from neutrophils cultured in the presence of 100 U/mL TNF-α (20 minutes at 37°C) were prepared and analyzed in EMSA. Before addition of the labeled NF-κB oligonucleotide probe to the binding mixture, extracts were incubated with a 10-, 25- or 50-fold molar excess of unlabeled NF-κB oligonucleotide (“cold”), or with an excess of unlabeled oligonucleotide containing mutated NF-κB binding sites (“mut”). The amount of extract used corresponded to 510,000 cell equivalents (approximately 16 μg of protein). (D) Neutrophils were cultured in the presence of LPS as described in (A), and nuclear extracts were analyzed in EMSA. Before addition of the labeled NF-κB oligonucleotide probe, extracts were incubated with antisera raised against p50 (no. 1141), p52 (no. 1267), c-Rel (no. 1136), the C-terminal region of p65/RelA (“C65”, no. 1226), the N-terminal region of p65 (“N65”, no. 1207), or without antisera (“-”). As a control, binding mixtures that only contained one of the antisera (in this case the anti-c-Rel, “Ab”), or normal rabbit serum (“NRS”), were routinely included on the gels. The amount of extract used corresponded to 960,000 cell equivalents, representing about 4.8 μg of protein. Supershifted complexes (“SS”) are indicated, as well as nonspecific bands (“NS”) present either in normal rabbit serum or in the control antiserum. Each of the experiments depicted in this figure is representative of at least four.
To ascertain the specificity of the aforementioned NF-κB DNA binding activities, competition experiments were performed using whole-cell extracts from LPS- or TNF-α-activated neutrophils. As shown in Fig 6C, the slower-migrating NF-κB DNA binding activity (complex A) was specific, as its detection was abolished in the presence of an excess of unlabeled NF-κB probe, whereas it was unaffected by competition with a 25-fold excess of unlabeled mutant oligonucleotide. The faster-migrating doublet present in the same extracts (complex B) was similarly affected by the competitor oligonucleotides, thereby showing specificity in its interaction with our labeled NF-κB probe. On the contrary, the fastest-migrating DNA binding activity (complex C) appeared to be nonspecific, as competitor oligonucleotides did not significantly impede its detection (Fig 6C). In similar experiments, the inclusion of rh IκB-α protein in the binding mixtures was found to dose-dependently displace the slower-migrating band (complex A), whereas the detection of the faster-migrating complexes (B and C) was not noticeably affected (data not shown). In an attempt to determine the subunit composition of the specific NF-κB complexes, supershift experiments were performed using nuclear extracts from LPS-activated neutrophils. Figure 6D shows that the migration of the inducible NF-κB band (complex A) was further retarded following coincubation of the extracts with anti-p50 or anti-RelA antisera, whereas an anti–c-Rel antiserum weakly, but consistently, impaired the detection of the complex. In contrast, the use of an anti-p52 antiserum invariably failed to exert a significant effect, consistent with our finding that the p52 protein is undetectable in human neutrophils. Identical results were obtained using either whole-cell extracts of LPS- or TNF-α–treated neutrophils, or DOC-treated whole-cell extracts from resting neutrophils (data not shown). Thus, it appears that the inducible NF-κB complex primarily consists of p50/p65 heterodimers, but that it may also contain minor amounts of c-Rel-containing complexes. Finally, it is noteworthy that the faster-migrating NF-κB DNA binding activities present in neutrophil whole-cell and cytoplasmic extracts (complexes B and C) were unaffected by any of the antisera (Fig 6D and data not shown). Similar bands were also weakly detected in monocyte or Jurkat extracts; in contrast to neutrophil extracts, these faster-migrating complexes were much less abundant than complex A, but were equally unaffected by our antisera.
In view of the fact that the nuclear translocation of NF-κB/Rel proteins followed different time courses depending on the stimulus, we next investigated the kinetics of NF-κB activation in neutrophils stimulated with various agonists. As shown in Fig 7A, the effect of LPS towards nuclear NF-κB activation was rapid and transient, as it was clearly detectable within 10 minutes, reached a maximum between 20 and 30 minutes, and had returned to near-basal levels by 60 to 90 minutes. A similar time course of NF-κB activation was observed when neutrophils were exposed to 100 U/mL TNF-α or 10 ng/mL IL-1β (Fig 7B), although the latter cytokine proved to be significantly less potent than LPS or TNF-α in inducing this response, in agreement with our immunoblot data. In contrast, stimulation of neutrophils with chemoattractants such as fMLP (Fig 7A) resulted in a delayed activation of NF-κB, insofar as it required between 30 and 40 minutes to be evident, depending on the donor; the magnitude of this response was also somewhat modest, relative to that observed following LPS or TNF-α stimulation. Another difference between the effect of fMLP and that of LPS or TNF-α is that in response to the latter agonists, a second increase in NF-κB DNA binding activity was sometimes detected after 90 minutes of stimulation (data not shown). We also examined the effect of a wide range of other neutrophil stimuli towards nuclear NF-κB activation. Figure 7B depicts the comparative ability of some of these agonists to activate NF-κB in neutrophils, and the overall results of these experiments are summarized in Table 2.
Effect of LPS, TNF-α, and fMLP towards NF-κB activation in human neutrophils. (A) Cells (5 × 106/mL) were cultured at 37°C in the presence or absence of 1 μg/mL LPS, 100 U/mL TNF-α, or 10 nmol/L fMLP for the indicated times (in minutes), and nuclear extracts were prepared and analyzed in EMSA. The amount of nuclear extract used in the binding reactions corresponded to 106 cell equivalents (upper panel) and 1.5 × 106 cell equivalents (lower panel), representing approximately 4.5 and 7.0 μg of protein, respectively. (B) Neutrophils were cultured in the presence or absence of 100 U/mL TNF-α, 10 ng/mL IL-1β, 10 nmol/L fMLP, 10 ng/mL PMA, or 10 nmol/L LTB4 as described in (A) for the indicated times (in minutes). Nuclear extracts were then prepared and analyzed in EMSA. The amount of nuclear extract used corresponded to approximately 750,000 cell equivalents. Each of the experiments depicted in this figure is representative of at least four.
Effect of LPS, TNF-α, and fMLP towards NF-κB activation in human neutrophils. (A) Cells (5 × 106/mL) were cultured at 37°C in the presence or absence of 1 μg/mL LPS, 100 U/mL TNF-α, or 10 nmol/L fMLP for the indicated times (in minutes), and nuclear extracts were prepared and analyzed in EMSA. The amount of nuclear extract used in the binding reactions corresponded to 106 cell equivalents (upper panel) and 1.5 × 106 cell equivalents (lower panel), representing approximately 4.5 and 7.0 μg of protein, respectively. (B) Neutrophils were cultured in the presence or absence of 100 U/mL TNF-α, 10 ng/mL IL-1β, 10 nmol/L fMLP, 10 ng/mL PMA, or 10 nmol/L LTB4 as described in (A) for the indicated times (in minutes). Nuclear extracts were then prepared and analyzed in EMSA. The amount of nuclear extract used corresponded to approximately 750,000 cell equivalents. Each of the experiments depicted in this figure is representative of at least four.
We finally performed control experiments to ensure that the inducible NF-κB DNA binding activity (complex A) detected in our neutrophil suspensions did not reflect the presence of contaminating monocytes. The rationale for these experiments is that although the proportion of contaminating monocytes in our neutrophil preparations is marginal (ie, less than 0.5%), monocytes were nevertheless found to express substantially more NF-κB/Rel proteins than neutrophils on an individual cell basis (Fig 1A). Neutrophil and monocyte-enriched suspensions from the same donor were, therefore, cultured in the presence or absence of 1 μg/mL LPS for 20 minutes at 37°C before EMSA analysis of the resulting whole-cell extracts. The 20-minute time point was selected because it allows for maximal NF-κB activation to occur in both cell types. Figure 8 shows that a similar extent of NF-κB activation (complex A) was observed using approximately three times less monocytes than autologous neutrophils. Thus, it appears that while monocytes have a markedly superior ability to activate NF-κB on a per-cell basis, a monocyte contamination of at least 25% would be required to account for the detection of the inducible NF-κB DNA binding activity observed in our neutrophil preparations. Consistent with these results is that when monocyte whole-cell extracts representing 100 times less cells than the corresponding neutrophil extracts (ie, to mimic the equivalent of 1% contaminating monocytes) were analyzed in EMSA, the monocyte extracts consistently failed to yield any detectable NF-κB signal (data not shown). These experiments clearly show that the minimal level of contaminating monocytes present in our neutrophil suspensions cannot reasonably account for the NF-κB DNA binding activities detected in our neutrophil extracts. Further support for this conclusion is that the p52 and p100 proteins were readily detected by immunoblot using 106 monocytes (Fig 1A), whereas they were undetectable using four times more neutrophils, and that IL-6 mRNA, which is indicative of monocyte contamination,2 39 was undetectable in our neutrophil preparations, as determined by Northern blot.
Comparative ability of neutrophils and autologous monocytes to activate NF-κB in response to LPS. Cells (5 × 106/mL) were cultured in the presence or absence ( — ) of 1 μg/mL LPS for 20 minutes at 37°C, and the resulting whole-cell extracts were analyzed in EMSA. The amount of extract used corresponded to 640,000 cell equivalents (neutrophils) or 190,000 cell equivalents (monocytes). This experiment is representative of three. The various DNA-binding complexes are indicated by the letters A, B, and C; pmn, polymorphonuclear neutrophils; mono, monocyte-enriched suspensions.
Comparative ability of neutrophils and autologous monocytes to activate NF-κB in response to LPS. Cells (5 × 106/mL) were cultured in the presence or absence ( — ) of 1 μg/mL LPS for 20 minutes at 37°C, and the resulting whole-cell extracts were analyzed in EMSA. The amount of extract used corresponded to 640,000 cell equivalents (neutrophils) or 190,000 cell equivalents (monocytes). This experiment is representative of three. The various DNA-binding complexes are indicated by the letters A, B, and C; pmn, polymorphonuclear neutrophils; mono, monocyte-enriched suspensions.
DISCUSSION
In this study, we report that among Rel family proteins, p50/NFκB1 (and its precursor, p105), as well as p65/RelA and c-Rel, are present in human neutrophils, albeit in smaller quantities than in autologous monocytes, while p52/NFκB2 (or its precursor, p100) was only detected in the latter cell type. It is tempting to speculate that this greater abundance of Rel family proteins in monocytes, in conjunction with the markedly superior extent to which monocytes (respective to neutrophils) activate NF-κB in response to stimuli such as LPS, might provide a partial explanation for the numerous observations in the literature showing that neutrophils have a much lower transcriptional activity than monocytes on an individual cell basis (reviewed in Cassatella1 ). In both cell types, IκB-α was strictly cytoplasmic, and accordingly, immunoreactive NF-κB/Rel proteins were principally recovered in the cytoplasmic fractions of resting cells. This is in keeping with the ability of IκB-α to directly interact with either p50, p65/RelA, or c-Rel,27,45 and thereby keep them sequestered in the cytoplasm. Coimmunoprecipitation experiments not only confirmed that in neutrophils, IκB-α is physically associated with protein complexes containing p50, p65, or c-Rel, but also showed that some of these cytosolic complexes could be identified as p50/c-Rel, p50/p65, and p65/c-Rel heterodimers. Despite their predominantly cytoplasmic localization, detectable amounts of NF-κB/Rel proteins were consistently observed in nuclear fractions of unstimulated neutrophils and monocytes, and this was not attributable to a cytosolic contamination of the nuclei. In keeping with these data, a constitutive nuclear NF-κB DNA binding activity was also detectable in both cell types. In neutrophils, this constitutive activity was found to comprise both p65/RelA and p50, and possibly some c-Rel–containing complexes, as well (our unpublished data). Similarly, p65/RelA was reported to be present in nuclear extracts of unstimulated human monocytic THP-1 cells,46 and NF-κB DNA binding activities containing p50 were detected in nuclear extracts of unstimulated peripheral blood monocytes and of various monocytic cell lines.47-49 Although the significance of these observations is unclear, it has been recently proposed that constitutive nuclear NF-κB might contribute to the constitutive expression of transcripts encoding κB-dependent genes.47 This suggestion stemmed from the finding that cycloheximide treatment of resting Mono Mac 6 monocytic cells increased both the constitutive nuclear NF-κB activity and the steady-state level of TNF-α mRNA.47 In agreement with these data, we observed that in unstimulated neutrophils, cycloheximide not only increased the amount of nuclear-associated NF-κB/Rel proteins (albeit without affecting the total cellular pool of these proteins), but also superinduced the steady-state level of IκB-α mRNA (unpublished data, January 1996). Nevertheless, clear evidence for the involvement of constitutive nuclear NF-κB in the basal expression of κB-dependent genes still awaits a more direct demonstration.
Among a wide range of neutrophil agonists, TNF-α, LPS, fMLP, and PMA were found to efficiently induce both the nuclear accumulation of NF-κB/Rel proteins and the concomitant degradation of cytoplasmic IκB-α, whereas IL-1β, LTB4 , and PAF proved to be weaker stimuli at the concentrations tested. Moreover, the onset of both processes was paralleled by the activation of nuclear NF-κB DNA binding activity. In contrast, GM-CSF, G-CSF, interferon-α (IFN-α), IFN-γ, IL-8, and IL-10 exerted no detectable effect on any of these responses (up to 120 minutes). From a general standpoint, these observations are in good agreement with the reported action of the same stimuli towards the activation of the NF-κB pathway in other cell types (reviewed in Baeuerle and Henkel18 and Siebenlist et al19 ). In the particular case of IL-1β, it must be stressed that although it is a potent inducer of Rel protein nuclear translocation, NF-κB DNA binding activity, and κB-dependent gene expression in many cell types, its stimulatory action towards cytokine and chemokine gene expression in neutrophils is rather limited. For instance, IL-1β is less potent than LPS or TNF-α in inducing its own gene expression4 or that of IL-8,34 in keeping with our present data concerning the moderate ability of IL-1β (compared with TNF-α or LPS) to mobilize NF-κB/Rel proteins to the nucleus and to activate NF-κB. Conversely, the nuclear mobilization of NF-κB/Rel proteins (in particular, that of p65/RelA and c-Rel) was also induced by fMLP in neutrophils, resulting in an increase in nuclear NF-κB DNA binding activity, even though fMLP has heretofore not been described as an inducer of NF-κB activation. Nevertheless, isolated studies had already provided indirect evidence that in granulocytes, fMLP may exert some of its actions by acting through the NF-κB pathway. In HL-60 cells differentiated along the granulocytic lineage, treatment with antisense oligonucleotides to p65/RelA reduced RelA expression and diminished the ability of fMLP to upregulate CD11b expression without affecting its ability to induce a respiratory burst.50 Similarly, exposure of neutrophils to FK506, a drug that hinders NF-κB activation in T cells,51 partially inhibited the fMLP-elicited synthesis of cellular proteins, despite a normal increase in intracellular calcium.52 These observations are, therefore, in good agreement with the fMLP-induced redistribution of Rel family proteins and NF-κB activation reported herein.
In neutrophils, the stimuli that promoted the nuclear accumulation of Rel family proteins and concomitant NF-κB activation could be subdivided into two general categories: those whose action was rapid and slower-acting ones. The effect of the former (TNF-α and LPS, and on a much smaller scale, IL-1β) was already evident by 10 minutes and had reached a maximum by 30 minutes. In contrast, the latter agonists (fMLP and PMA, and to a lesser extent, PAF and LTB4 ) required at least 30 minutes to exert a similar action, a maximal effect being usually observed between 45 and 60 minutes. This delayed induction is not likely to reflect a requirement for endogenously produced mediators such as TNF-α or IL-1β, as fMLP lacks the ability to induce their release from neutrophils (unpublished data, May 1992). Similarly, neither the nuclear accumulation of NF-κB/Rel proteins, nor the activation of NF-κB, could be prevented by a prior treatment of the cells with cycloheximide (unpublished data, December 1995), indicating that this response cannot be attributed to newly synthesized proteic factors. As in most other cell types studied to date, the nuclear translocation of Rel family proteins and concurrent activation of NF-κB in neutrophils appeared to depend on the degradation of IκB-α, as the onset of all three processes occurred in close parallel, regardless of the type of agonist used. More direct evidence is the finding that AEBSF (a protease inhibitor) not only blocked the inducible degradation of IκB-α, as already reported,53 but also prevented both the nuclear accumulation of Rel family proteins and NF-κB activation under the same conditions (unpublished data, January 1996). Therefore, the differences observed between fast-acting and slower-acting stimuli must reflect a differential ability to activate cellular processes that lead to IκB-α degradation, such as those that bring about its phosphorylation. The central role of the events controlling IκB-α expression within the context of the NF-κB pathway is further illustrated by the fact that regardless of the stimulus used, reexpression of the inhibitor protein correlated well with the termination of nuclear NF-κB DNA binding activity in neutrophils. It must be recalled, however, that nuclear levels of Rel family proteins remained elevated even when NF-κB DNA binding activity was no longer detected. This apparent contradiction might find its explanation in light of a recent study, which showed that in HeLa S3 cells briefly exposed to TNF, and then washed free of the stimulus, the termination of NF-κB activation correlated with the detection in nuclear extracts of a fraction of the de novo–synthesized IκB-α, which was physically associated with RelA- or p50-containing complexes.26 Under these conditions, nuclear levels of p50 eventually decreased with time. In contrast, when HeLa S3 cells were continuously exposed to TNF (as in the present study), IκB-α was only weakly detected in nuclear extracts, despite the fact that considerable amounts of extract were processed for immunoblot analysis.26 Moreover, the loss of p50 from nuclear fractions of TNF-treated cells was not evident under these conditions,26 similar to the present observations. Thus, it is conceivable that in neutrophils that are continuously exposed to a given stimulus, a small amount of de novo-synthesized IκB-α might localize to the nucleus, which could suffice to prevent NF-κB binding, yet without substantial changes in nuclear levels of Rel family proteins. Alternatively, the loss of NF-κB DNA binding activity observed at late time points in stimulated neutrophils might reflect changes in the phosphorylation status of nuclear NF-κB/Rel proteins. In this regard, it has been reported that p65/RelA becomes transiently phosphorylated following TNF-α stimulation of astrocytes, and that in TNF-stimulated HeLa cells, RelA phosphorylation occurs in parrallel with that of IκB-α and p105.54,55 Similarly, phorbol ester stimulation of Jurkat T cells has been shown to result in the phosphorylation of both p105 and p50.56 More importantly, these studies have demonstrated that the phosphorylated forms of RelA and p50 both confer a considerably enhanced DNA-binding activity to NF-κB dimers, relative to their unphosphorylated counterparts.54 56 Thus, it can be envisaged that in stimulated neutrophils, the nuclear accumulation of Rel proteins might be paralleled by their phosphorylation, and that the termination of NF-κB activation might partially reflect a subsequent dephosphorylation, despite elevated nuclear levels of NF-κB/Rel proteins. Similarly, the significant amounts of Rel proteins found in the nuclei of resting neutrophils might exhibit low DNA-binding activity as a result of their being unphosphorylated.
We finally demonstrated that in neutrophils, the degradation of the IκB-α protein and concomitant nuclear activation of NF-κB were accompanied by a marked accumulation of IκB-α mRNA transcripts, resulting in the reexpression of the protein itself, through de novo synthesis. This inducible accumulation of IκB-α mRNA was prevented by actinomycin D pretreatment, indicating the involvement of transcriptional events, and was protein synthesis-independent, in that it was not inhibited by cycloheximide pretreatment. Together, these observations raise the possibility that in neutrophils, the inducible accumulation of IκB-α transcripts might rely on the activation of preexisting transcription factors. In view of the known requirement for NF-κB complexes to bind their cognate sites within the IκB-α gene promoter to enhance gene expression,22-25 it can be reasonably argued that these transcription factors must include members of the NF-κB family. In keeping with this assertion is that the same stimuli, which were found to upregulate IκB-α gene expression in neutrophils, were also found to promote NF-κB activation. In a broader context, these stimuli also have the ability to induce the expression of other κB-dependent genes in neutrophils, in particular those encoding cytokines and chemokines.2-8,17,33 As in the case of IκB-α, the nuclear mobilization of Rel family proteins occurring in response to these stimuli is likely to result in increased gene expression, inasmuch as the binding of Rel protein dimers to the κB and κB-like motifs located in the 5′ regulatory region of the genes encoding TNF-α, IL-1β, IL-8, and MIP-1α is sufficient to confer transcriptional inducibility,12-15 and is required for the full activation of the IL-1ra gene promoter.16 In conclusion, the coordinate translocation of NF-κB/Rel proteins to the nuclear compartment, where they can bind as homo- or heterodimers to the κB or κB-like motifs located in the promoter region of κB-dependent genes, could constitute one of the mechanisms whereby the expression of these genes is enhanced on neutrophil stimulation. Studies are in progress to further define the potential involvement of NF-κB activation in the expression of selected genes in human neutrophils.
ACKOWLEDGEMENT
We are particularly indebted to Dr Nancy Rice for generously providing many antisera. We also thank Federica Calzetti for her reliable assistance in performing Northern blots.
Supported by grants to M.A.C. from the Consiglio Nazionale delle Ricerche (progetto strategico citochine 95.02809.ST74), Roma, from the Istituto Superiore della Sanità (VIII progetto AIDS 9304-33), Roma, and from the Associazione Italiana per la Ricerca contro il Cancro, Milano. P.P.M. is a postdoctoral Fellow of the Medical Research Council of Canada.
Address reprint requests to Marco A. Cassatella, MD, Istituto di Patologia Generale, Università di Verona, Strada Le Grazie 4, 37134 Verona, Italy.