Abstract
We present two novel alleles of the anion-exchanger 1 (AE1) gene, allele Coimbra and allele Mondego. Allele Coimbra (V488M, GTG → ATG) affects a conserved position in the putative second ectoplasmic loop of erythrocyte band 3. In 15 simple heterozygotes, it yielded a mild form of hereditary spherocytosis (HS) with band 3 deficiency (−20% ± 2%) and a reduced number of 4,4′-diisothiocyano-1,2-diphenylethane-2,2′-disulfonate (H2DIDS) binding sites (−35%). However, two additional heterozygotes presented with an aggravated HS and a more pronounced reduction of band 3 (−40%) and of H2DIDS binding sites (−48%). They carried, in trans to allele Coimbra, allele Mondego, defined by two mutations: E40K, GAG → AAG, the known mutation Montefiore, and P147S, CCT → TCT, a novel mutation, both located in the cytoplasmic domain of band 3. Allele Mondego itself resulted in no clinical or hematologic HS signs in the simple heterozygous state. Yet it yielded a slight decrease in band 3 (−6% to −12%) and in the number of H2DIDS binding sites (−19%). Thus, the more pronounced decrease in band 3 in the two compound heterozygotes derived from the additive effects of two unequally expressed AE1 alleles, resulting in a more severe clinical picture.
BAND 3, or anion-exchanger 1 (AE1), is the most abundant protein (1,200,000 monomers) of the red blood cell (RBC) membrane (for review, see Tanner1 ). It is composed of 911 amino acids.2,3 The first 403 residues constitute the cytoplasmic domain that binds a number of proteins, including ankyrin and protein 4.2. Residues 404 to 883 account for the membrane domains, and harbor 14 membrane-spanning segments (TM). The oligomeric forms of band 3 in situ are mainly tetramers and dimers.4-6 The AE1 gene encoding band 3 maps to 17q21-qter.7 It extends over 20 kb, approximately, and consists of 20 exons.8 9
Hereditary spherocytosis (HS) is a common hereditary hemolytic anemia in which the spheroidal, osmotically fragile RBCs are prematurely destroyed in the spleen (for review, see Lux and Palek10 ). A subset of HS (10% to 20%) is associated with mild to moderate clinical signs, band 3 deficiency (20% to 40%), a proportional secondary decrease in protein 4.2,11-15 and a dominant mode of inheritance. This subset has been found to be associated with highly heterogeneous mutations of the AE1 gene.12-21 Another subset of rare HS is characterized by a conspicuous deficiency of protein 4.2; no band 3 deficiency was mentioned.22-25 The corresponding mutations located in the cytoplasmic domain of band 3 presumably alter the protein 4.2 binding site.
HS associated with band 3 deficiency is usually homogeneous with regard to the clinical and biochemical picture within a given family. However, in certain families, some individuals are sicker than others, and this, at least in some cases, seems correlated with a more pronounced decrease in band 3. We recently showed that the mild HS associated with the AE1 allele Lyon (carrying a nonsense mutation) was aggravated when AE1 allele Genas occurred in trans.13 Allele Genas bears a nucleotide substitution in the 5′ untranslated region (UTR) concomitant with a partial mRNA deficiency. In the simple heterozygous state, allele Genas is symptomless, although it results in a slight reduction in band 3 (∼−6%).
We report two novel abnormal alleles of the AE1 gene generating band 3 deficiency to different degrees. Heterozygous allele Coimbra (V488M) generated symptomatic HS and a frank decrease in band 3. Heterozygous allele Mondego (E40K, P147S) produced a slight deficiency in band 3 but remained symptomless. In the compound heterozygous state, HS was aggravated and band 3 deficiency more pronounced.
SUBJECTS AND METHODS
Case Reports
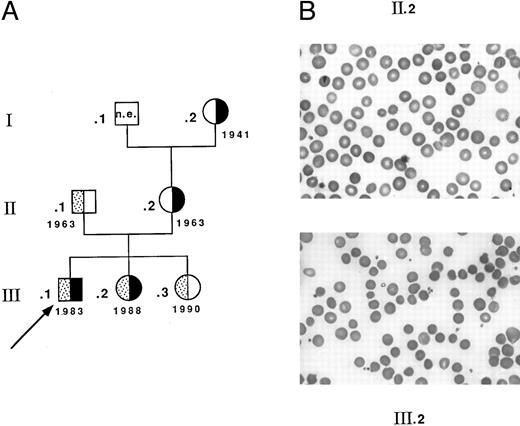
Seventeen heterozygotes carrying allele Coimbra were identified among 33 individuals investigated from related Portuguese families CL and MG. Fifteen simple heterozygotes (13 from family CL (complete genealogic tree not shown) and two from family MG, I.2 and II.2, Fig 1) had a mild dominant HS: slight sclerotic icterus, inconstantly palpable spleen, usually compensated anemia, spherocytes (with some pincered RBCs), and reduced osmotic resistance.26 27 Four patients had been splenectomized as adults. In contrast, two children from family MG, carrying allele Coimbra, presented with a more severe picture of constantly uncompensated anemia (Fig 1). Child III.1, the proband, had neonatal jaundice and severe hemolytic anemia in the first months of life (hemoglobin [Hb] 51 g/L, RBC 1.9 T/L, reticulocytes 14%, and bilirubin 35 μmol/L). During the following years, Hb fluctuated between 80 and 85 g/L, but was lower during febrile episodes. Spherocytes were more conspicuous (Fig 1) and the osmotic resistance (not shown) was much more reduced. Splenectomy performed at 8 years of age was efficient (Hb 131 g/L, RBC 4.0 T/L, reticulocytes 4%, and bilirubin 13 μmol/L). Child III.2 had a comparable clinical presentation and responded identically to splenectomy at 6 years of age. Both children inherited allele Coimbra from their mother (II.2) and allele Mondego from their father (II.1). The latter was symptomless and had normal RBC data and normal osmotic resistance (Hb 156 g/L, RBC 4.5 T/L, reticulocytes 1.8%, and bilirubin 16 μmol/L). Child III.3 was also heterozygous for allele Mondego and, as did her father, remained symptomless.
Family MG. (A) Genealogic tree and birth dates: ▪, allele Coimbra; ▧ allele Mondego; ▧, proband. (B) Blood smears. Spherocytosis is more pronounced in child III.2 (and in child III.1, not shown) than in the mother II.2.
Family MG. (A) Genealogic tree and birth dates: ▪, allele Coimbra; ▧ allele Mondego; ▧, proband. (B) Blood smears. Spherocytosis is more pronounced in child III.2 (and in child III.1, not shown) than in the mother II.2.
Methods
Studies on erythrocyte membrane proteins.RBC membrane proteins were analyzed by polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS-PAGE) using a 3.5% to 17% exponential gradient,28 and scanned at 570 nm using a System II Densitometer (Sebia, Issy-les-Moulineaux, France). The band 3 to spectrin, band 3 to protein 4.1, and protein 4.2 to protein 4.1 ratios were calculated using Fairbanks gel.28 Western blots were performed using mouse monoclonal anti–human band 3 antibodies (Sigma, St Louis, MO). Erythrocytes were fractionated according to density using a Percoll gradient.29,30 To determine if anion transport was reduced in proportion to band 3 deficiency, 4,4′-diisothiocyano-1,2-diphenylethane-2,2′-disulfonate (H2DIDS) binding sites and sulfate flux were determined using H2DIDS as described by Schofield et al.31
Based on the presence of mutation Montefiore (lysine at position 40) in band 3 Mondego, we used trypsin digestion (trypsin/substrate 1/500 to 1/50; incubation for 15 minutes to 1.5 hours at 4°C) of stripped vesicles (pH 12.0) according to the method of Rybicki et al23 to assess the presence of band 3 Mondego. Peptide maps were analyzed using 12% SDS-PAGE32 and densitometric scanning (570 nm). Band 3 Mondego generated a 16-kD truncated N-terminal fragment. Normal band 3 and band 3 Coimbra produced a 22-kD normal N-terminal fragment. The 16-kD/(22-kD + 16-kD) ratio was determined to estimate the ratio for band 3 Mondego to total band 3. Assuming that the staining was roughly proportional to the polypeptide length, we applied the multiplication factor, 22/16 = 1.37, to the surface value of the 16-kD band.
Studies on band 3 cDNA and AE1 gene.Reticulocyte RNA was prepared from peripheral blood as polysomal precipitates and extracted with phenol, chloroform, and isoamyl alcohol.33 Reverse transcription (RT) was performed as previously described,34 followed by polymerase chain reaction (PCR). Only nucleotide primer sequences not presented elsewhere12,13 will be described here and numbered from the ATG initiation codon. Primers E/F and G/H12,13 were used to cover the entire transmembrane coding sequence. The products were digested with BanII and HinfI + BstUI, respectively, to generate subfragments suitable for the search for single-strand conformation polymorphisms (SSCPs).12 35
Direct nucleotide sequencing was performed with asymmetrical RT-PCR (primers E/F, primer F in excess) using primer X (5′AGTGTCGAGCTGCTGATCT, sense; nucleotide [nt] 1308 to nt 1327) as previously described.34 Determination of the entire coding sequences and parts of the 5′-and 3′-UTR of cloned band 3 Coimbra and band 3 Mondego cDNAs from a compound heterozygote (MG III.2) was accomplished. Coimbra and Mondego clones were differentiated based on the NlaIII site (created by mutation Coimbra) before sequencing. We cloned PCR fragments (primer A12,13 and primer 5′GAGAAGATCTCCTGGGTATAG, antisense, nt 1574 to nt 1554) into the pCRII vector (TA cloning Kit; Invitrogen, San Diego, CA). We cloned PCR fragments (primers E/H with the additional sequence GCGAATTC at the 5′ end) into the pUC18 EcoRI/BAP plasmid (Pharmacia, Piscataway, NJ).
Genomic DNA was obtained from leukocytes.36 In view of preliminary linkage studies, we amplified a DNA fragment carrying the PstI polymorphism.37 We used primers 5′GGGTGTTCCCTGGCATAG (sense, intron 2) and 5′TCTCCCCCAGCCTGAAGCT (antisense, intron 4), and digested the product with PstI endonuclease. Exon 13 mutation (Coimbra) was screened using NlaIII digestion following genomic DNA amplification with primer X (see above) and primer 5′TGATCTCGGGTGATCCACCT (antisense, intron 13). Exon 4 mutation (mutation Montefiore within allele Mondego) created a StyI site and was screened following genomic DNA amplification (primers L/M13). Exon 6 mutation (P147S within allele Mondego) abolished a Bsu36I site and was screened following genomic DNA amplification (primers N/O13).
We estimated the amount of normal and mutant mRNA using RT-PCR. The conditions under which the amount of amplified DNA and the incubation time were linearly related were preliminarily determined for each following PCR. Then, samples with mutation Coimbra were submitted to 27 cycles (primers E/F ) and digested with NlaIII. Samples with allele Mondego were submitted to 23 cycles (primer A and antisense primer 5′CTAGGCCCTTGTAGAAGCTG, nt 1087 to nt 1068) and digested with Bsu36I. Bands were measured densitometrically following electrophoresis and ethidium bromide staining.
RESULTS
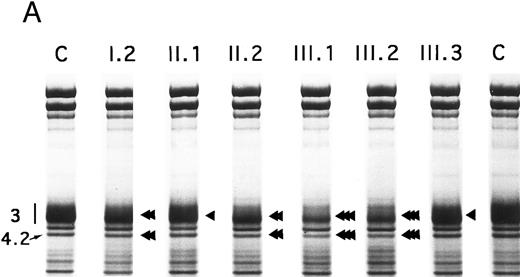
Proteins, H2DIDS binding sites, and sulfate flux.Using SDS-PAGE,28 the decrease in band 3 was similar in the 13 normal/Coimbra (HS) patients of family CL (band 3/spectrin, −20% ± 2%; band 3/protein 4.1, −25% ± 3%; not shown). A similar band 3 reduction was observed for normal/Coimbra patients I.2 and II.2 of family MG (band 3/spectrin, ∼−18%; band 3/protein 4.1, ∼−27%; Table 1 and Fig 2). There was also a proportional reduction in protein 4.2 (protein 4.2/protein 4.1, −18% ± 3% in the 13 patients of family CL, not shown; −17% and −20% in patients I.2 and II.2 of family MG, respectively; Fig 2).
SDS-PAGE of membrane proteins (A) and densitometric scanning (B) in members of family MG. C, controls. II.1 and III.3 (slight, ◂), I.2 and II.2 (moderate, ◂◂) and III.1 and III.2 (pronounced ◂◂◂), reductions of band 3 and protein 4.2.
SDS-PAGE of membrane proteins (A) and densitometric scanning (B) in members of family MG. C, controls. II.1 and III.3 (slight, ◂), I.2 and II.2 (moderate, ◂◂) and III.1 and III.2 (pronounced ◂◂◂), reductions of band 3 and protein 4.2.
In Coimbra/Mondego individuals III.1 and III.2 of family MG (presenting with an aggravated clinical picture), the reductions were more pronounced for band 3 (∼−40%; Table 1 and Fig 2) and protein 4.2 (∼−37%; Fig 2). (Although the amounts of band 3 and protein 4.2 were measured before splenectomy in child III.2 and after splenectomy in child III.1, the values were similar.)
There was a slight decrease in band 3 in normal/Mondego individuals II.1 and III.3 in family MG (−6% for band 3/spectrin or −12% for band 3/protein 4.1; Table 1 and Fig 2), but protein 4.2 was not reduced (Fig 2).
Minor bands of abnormal size were not observed with immunoblotting. The size and quantity of spectrin, ankyrin, protein 4.1, and actin were normal (not shown). The reduction of band 3 was similar in dense (reticulocyte-poor) and in light (reticulocyte-rich) fractions in normal/Coimbra, normal/Mondego, and Coimbra/Mondego individuals (not shown).
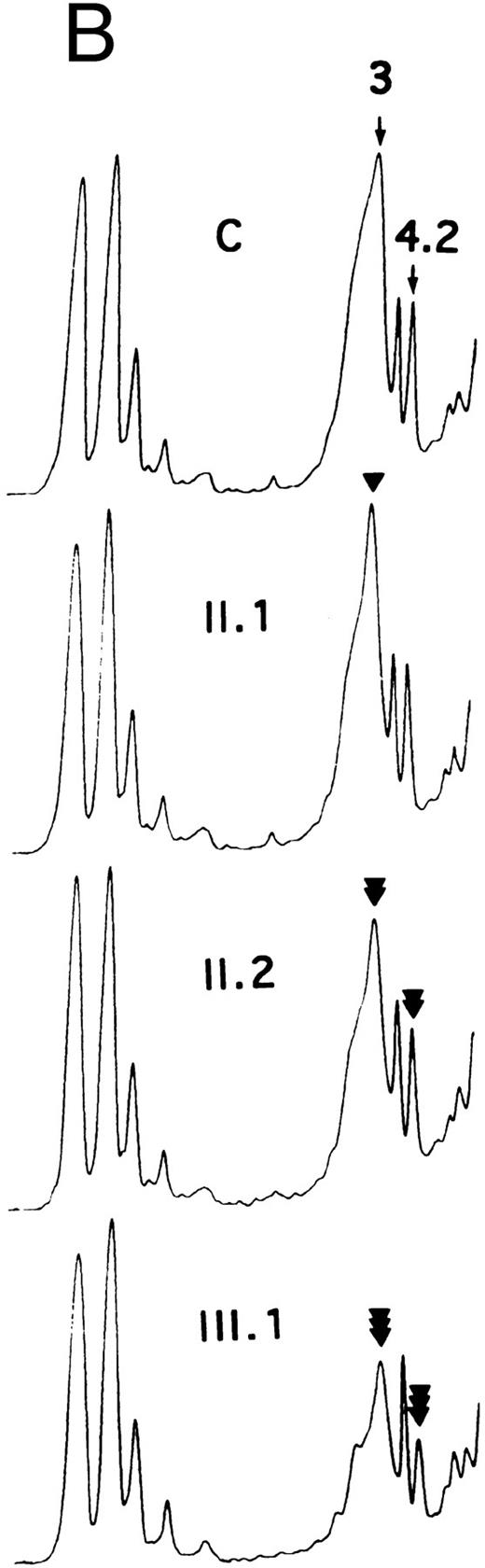
The number of H2DIDS binding sites and the rate of sulfate flux were significantly reduced in individuals II.1 and III.3 (Table 1). Individuals III.1 and III.2 exhibited a more dramatic reduction, whereas the reduction for individual II.2 was intermediate. The decrease in the number of H2DIDS binding sites and the rate of sulfate flux reflected the lower amount of band 3. However, the small discrepancy between these values has already been observed by others16 17 and reflects at least in part, the greater accuracy of the measurements of transport activity as compared with SDS-PAGE.
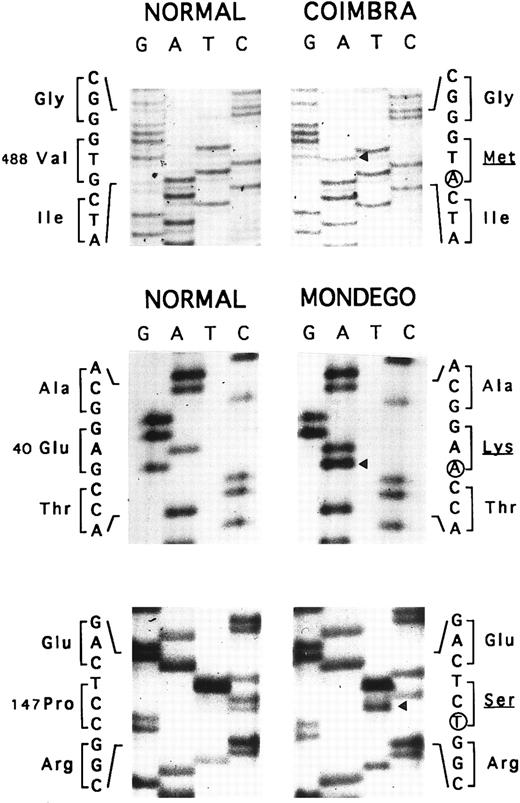
Allele Coimbra.In a preliminary approach, the PstI polymorphic site of the AE1 gene indicated that HS was associated with a PstI (+) allele (not shown). SSCP analysis of the membrane domain sequence in two patients showed a conformational change in the cDNA segment (328 nt) extending from nt 1368 to 1695 (not shown). Direct nt sequencing (Fig 3) disclosed the mutation, V488M (GTG → ATG, exon 13), defining allele Coimbra and occurring in the putative second ectoplasmic loop.2 3 A NlaIII site was created (+, 293- + 86-nt fragments; −, 379-nt fragment). The NlaIII site was present in the heterozygous state in 17 HS patients, including individuals I.2, II.2, III.1, and III.2 of family MG, and was absent in 16 normal individuals (not shown). The nt sequencing was completed to the entire coding sequence of cloned cDNA Coimbra and to parts of the 5′- and 3′-UTR 148 nt upstream from the initiator ATG and 79 nt following the stop TGA codon. No other changes were observed.
Nt sequencing. Allele Coimbra (◂, V488M, GTG → ATG), direct sequencing. Allele Mondego (◂, E40K, GAG → AAG and P147S, CCT → TCT), sequencing following cloning.
Nt sequencing. Allele Coimbra (◂, V488M, GTG → ATG), direct sequencing. Allele Mondego (◂, E40K, GAG → AAG and P147S, CCT → TCT), sequencing following cloning.
Allele Mondego.We assumed that the most affected children, III.1 and III.2, had inherited from their father II.1 a slightly deficient AE1 allele not clinically expressed in the simple heterozygous state. Band 3 cDNA from individual III.2 was cloned. We found clones with mutation Coimbra (see above) and others without this mutation. The complete coding sequences of the non-Coimbra cDNA and parts of the 5′- and 3′-UTR (as above) were determined. Two mutations in the cytoplasmic domain sequences were observed: E40K (GAG → AAG, exon 4), reported before as mutation Montefiore,23 and P147S (CCT → TCT, exon 6), a novel mutation (Fig 3). The double-mutated alleles were named Mondego. The mutation at position 40 created a StyI site (+, 185- + 181-nt fragments; −, 366-nt fragment), and that at position 147 abolished a Bsu361 site (+, 230- + 212-nt fragments; −, 442-nt fragment). Screening these sites revealed allele Mondego in individuals II.1, III.1, III.2, and III.3.
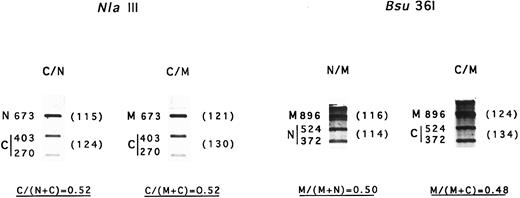
mRNA levels.Determination of mRNA content was performed with two independent RT-PCR and digestion procedures. The results from NlaIII digestion suggested that about 50% of the mRNA was Coimbra mRNA in both Coimbra simple heterozygotes and Coimbra/Mondego compound heterozygotes (Fig 4). Identical results were obtained based on Bsu36I digestion of mRNA Mondego (Fig 4). Thus, no transcriptional or posttranscriptional defects were evidenced.
Amount of mRNA Coimbra and mRNA Mondego. N, normal cDNA fragment; C, Coimbra cDNA fragment; M, Mondego cDNA fragment. Two different RT-PCR amplification products were used. They were digested with NlaIII (+, 403- + 270-nt bands; −, 673 nt) and Bsu36I (+, 524- + 372-nt; −, 896 nt), respectively, and electrophoresed. The fragments were stained with ethidium bromide and scanned. Parentheses contain the peak surface areas (arbitrary units).
Amount of mRNA Coimbra and mRNA Mondego. N, normal cDNA fragment; C, Coimbra cDNA fragment; M, Mondego cDNA fragment. Two different RT-PCR amplification products were used. They were digested with NlaIII (+, 403- + 270-nt bands; −, 673 nt) and Bsu36I (+, 524- + 372-nt; −, 896 nt), respectively, and electrophoresed. The fragments were stained with ethidium bromide and scanned. Parentheses contain the peak surface areas (arbitrary units).
Amount of band 3 Coimbra and band 3 Mondego in the membrane.Trypsin digestion showed that the concentration of band 3 Mondego was higher in Coimbra/Mondego compound heterozygotes than in simple Mondego heterozygotes (not shown). Using various enzyme/vesicle ratios and various incubation times, the maximal band 3 Mondego percentage (16-kD/16-kD + 22-kD) was about 70% in compound heterozygotes and 40% in simple heterozygotes. We cannot exclude the possibility that some band 3 Mondego remained undigested (22-kD fragment). Thus, band 3 Mondego was less well incorporated than normal band 3, and band 3 Coimbra was less well incorporated, if at all, than band 3 Mondego.
DISCUSSION
These results support the hypothesis that allele Coimbra is responsible for HS: (1) Allele Coimbra was present in 17 HS patients investigated and in none of 16 normal members from the two families; (2) The nt sequence of the whole coding segment and of substantial portions of the 5′- and 3′-UTR showed no other changes; and (3) Valine 488 was conserved in all known AE1 cDNA (five species) and AE2 cDNA (four species) and replaced by threonine in AE3 cDNA (three species).38-40 This degree of conservation is similar to that reported for different recent amino acid substitutions found in other HS cases with band 3 deficiency.15,16 20
In the membrane domain, 11 other substitutions of conserved residues have been described in association with band 3 deficiency.12,14-16,20. They were located in TM segments or adjacent to them in the inner or outer loops. Most of them (9 out of 11) were clustered in the C-terminal half of the membrane domain (TM9 to 14). The two remaining changes, G455E15 and R518C,20 were located at the end of TM2 and in the second inner loop, respectively. In one case, R760Q, Jarolim et al16 demonstrated that the mutant band 3 was not incorporated into the membrane. On the other hand, comparable mutations in the putative third ectoplasmic loop, P548L41 and V557M41,42 carrying the Rb and Wd blood group antigens, respectively, yielded no band 3 deficiency even though P548 is even better conserved than V488.38 A possible explanation is that the third ectoplasmic loop (putative positions 543 to 567) is longer than the second one (putative positions 480 to 490) and contains many variable amino acids. As a result, its greater plasticity would accommodate a change (P548L) that even involves a conserved position.
The more pronounced band 3 deficiency in the two sicker children resulted from the additive effects of a common AE1 allele causing HS with band 3 deficiency and a symptomless allele generating only a slight band 3 deficiency. In compound heterozygotes, the reductions of the band 3/protein 4.1 ratio (−41%) and of the number of H2DIDS binding sites (−48%) were equivalent to the sum of the corresponding deficiencies in simple heterozygotes − (12 + 27) = −39% and − (19 + 35) = −54%, respectively (Table 1). However, summing the reduction of the band 3/spectrin ratios in the two kinds of simple heterozygotes, − (6 + 18) = −24% did not equal the value in the compound heterozygotes, −40%, nor did the addition of sulfate flux rates, − (12.5 + 34) = −46.5% versus −61.5%. It is not possible to conclude whether these small discrepancies are significant.
The amino acid substitutions in the cytoplasmic domain of band 3 yield a small decrease of the variant band 3. The first mutation of allele Mondego (E40K) introduces a basic residue at a poorly conserved position within the acidic segment.38-40 This mutation, known as mutation Montefiore,23 has previously been reported to be associated (1) in the homozygous state with HS and a sharp decrease of protein 4.2, but not with band 3 deficiency; and (2) in the heterozygous state with no symptoms or any reduction of band 3 and protein 4.2. Mutation Montefiore has been encountered in the heterozygous state in a few other individuals,16,26 and one case investigated in this respect was shown not to be linked with the P147S mutation (S.E. Lux and S.W. Eber, personal communication, January 1997). Screening 57 normal unrelated Portuguese persons for this mutation, we found two heterozygotes, ie, an allele frequency of 0.02. A similar frequency was obtained by screening 23 unrelated Portuguese HS patients (1 out of 46 alleles tested; it is unlikely, though, that heterozygous mutation Montefiore per se accounted for HS in this particular case). These results are in agreement with those of Jarolim et al16 (2 out of 48 HS patients tested). Thus, mutation Montefiore occurs relatively frequently. Mutation P147S removes a helix-breaking residue at a conserved position (proline in four of five AE1 species (Ala in the trout) and in three of three AE3 species, and alanine in four of four AE2 species). It was not found in the 57 controls (especially in the two Montefiore heterozygous controls) or in the 23 HS patients (particularly in the Montefiore heterozygous patient). Mutation P147S seems to be a isolated mutation (allele frequency < 0.006, 0 out of 160 alleles tested).
Allele Mondego, like allele Genas,13 is clinically unexpressed in the heterozygous state. One must conceive that HS symptoms do not develop as long as the reduction in overall band 3 remains below an ill-defined but probably low threshold. Two changes in the cytoplasmic domain of band 3 have been shown to cause in the heterozygous state a clinically manifest HS with band 3 deficiency: a deletion of five amino acids (positions 117 to 121) and, more intriguingly, a mere A285D substitution.15
These results highlight the clinical importance of AE1 gene mutations that are clinically silent in the simple heterozygous state even though they yield a slight decrease in band 3. By an additive effect, they produce a further decrease in band 3 and a clinical worsening when they occur in trans to common AE1 mutations causing HS and band 3 deficiency in the simple heterozygous state.
ACKNOWLEDGMENT
We thank family CL and related branches and family MG for their kind cooperation; Dr L. Flores for performing most of the field studies; Dr R. Wilmotte for technical advice; Dr U. Rebelo, H. Almeida, and C. Vasseur for their precious help; and N. Connan for preparing the manuscript.
Supported by the Université Claude Bernard Lyon-I, the Centre National de la Recherche Scientifique (URA 1171 and PICS 221, a joint program with the Ministère des Affaires Etrangères, Paris, France, and with the Ministry of Education (Monbusho), Tokyo, Japan), the Institut National de la Santé et de la Recherche Médicale (CRE 930405), the Conseil Régional de la Région Rhône-Alpes, the Association Française contre les Myopathies, the Fondation pour la Recherche Médicale, the Association de Recherche contre le Cancer, the Programme Hospitalier de Recherche Clinique (1995-1997), and the Institut Pasteur de Lyon.
Presented in part at the 35th Annual Meeting of the American Society of Hematology, St Louis, MO, December 3-7, 1993 and published in Blood 82:4a, 1993 (suppl 1, abstr).
Address reprint requests to N. Alloisio, PhD, Génétique Moléculaire Humaine, CNRS UMR 5534, CGMC, Bâtiment 741, Université Lyon I, 43, boulevard du 11 Novembre 1918, 69622 Villeurbanne Cedex, France.