Abstract
Using a murine bone marrow transplantation model, we evaluated the long-term engraftment of retrovirally transduced bone marrow cells in nonmyeloablated hosts. Male bone marrow was stimulated in a cocktail of interleukin-3 (IL-3), IL-6, IL-11, and stem cell factor (SCF ) for 48 hours, then cocultured on the retroviral producer line MDR18.1 for an additional 24 hours. Functional transduction of hematopoietic progenitors was detected in vitro by reverse transcriptase-polymerase chain reaction (RT-PCR) amplification of multiple drug resistance 1 (MDR1) mRNA from high proliferative potential-colony forming cell (HPP-CFC) colonies. After retroviral transduction, male bone marrow cells were injected into nonablated female mice. Transplant recipients received three TAXOL (Bristol-Myers, Princeton, NJ) injections (10 mg/kg) over a 14-month period. Transplant recipient tissues were analyzed by Southern blot and fluorescence in situ hybridization for Y-chromosome–specific sequences and showed donor cell engraftment of approximately 9%. However, polymerase chain reaction amplification of DNAs from bone marrow, spleen, and peripheral blood showed no evidence of the transduced MDR1 gene. RT-PCR analysis of total bone marrow RNA showed that transcripts from the MDR1 gene were present in a fraction of the engrafted donor cells. These data show functional transfer of the MDR1 gene into nonmyeloablated murine hosts. However, the high rates of in vitro transduction into HPP-CFC, coupled with the low in vivo engraftment rate of donor cells containing the MDR1 gene, suggest that the majority of stem cells that incorporated the retroviral construct did not stably engraft in the host. Based on additional studies that indicate that ex vivo culture of bone marrow induces an engraftment defect concomitantly with progression of cells through S phase, we propose that the cell cycle transit required for proviral integration reduces or impairs the ability of transduced cells to stably engraft.
HEMATOPOIETIC PROGENITOR cells, because of their renewal capacity as well as multilineage contribution, are an ideal target for the delivery of gene therapy for both disease treatment and genetic “rescue.”1-3 Gene transfer into hematopoietic cells by retroviral transduction has proven partially successful giving low-level and often short-term expression of the transduced genes both in human and animal models. The delivery of hematopoietic progenitor cells transfected with a foreign gene into myeloablated hosts, which received either substantial chemotherapy or irradiation, leads to marrow reconstitution and variable degrees of gene expression.4-18
The capacity of hematopoietic progenitor cells to lead to substantial engraftment in normal nonmyeloablated hosts, gives rise to reasonable expectation that transduced progenitor cells may be capable of delivering new gene sequences in a clinical or therapeutic situation where transplant without myeloablation is desirable, such as in the case of genetic diseases or human immunodeficiency virus (HIV). A murine transplant model, which shows reproducible engraftment kinetics for normal bone marrow, has been well characterized and used extensively to establish the nature of the long-term repopulating progenitor cells.19-22 The ability to deliver progenitor cells into a normal host provides the opportunity to assess the potential of transduced (retrovirally transfected) progenitor cells to engraft in the nonmyeloablated environment. The therapeutic potential for delivery of genes to a nonablated host is relevant to gene therapy applications, and transferring a selectable marker would offer operational and therapeutic advantages.
The human MDR1 multiple drug resistance 1 (MDR1) gene produces a membrane protein that confers resistance to p170 pumped drugs in a variety of cell types including hematopoietic lineages.23 Transplant of MDR1-transduced bone marrow cells confers resistance to TAXOL (Bristol-Myers, Princeton, NJ) in irradiated W/WV mice.24 Demonstration of the MDR1 transgene after serial transplantation through several generations of lethally irradiated hosts receiving 5FU-treated transduced cells25 26 indicates that integration occurs in a very early progenitor. The expression of MDR1 in cells of the hematopoietic lineages should provide host resistance to a variety of chemotherapuetic agents. This protects the marrow cells from the effects of the drugs and may allow safe delivery of greater therapeutic doses. Overexpression of the MDR1 gene, along with another therapeutic agent in a dicistronic vector would also permit the positive selection of progenitor cells in vitro or in vivo, which may be of special benefit when transplanting transduced cells into a normal host.
In this report, using an MDR1 retroviral vector, we show a high rate of successful transduction of primitive hematopoietic cells in vitro by mRNA expression of the MDR1 transgene in hematopoietic progenitor cell-derived colonies (multifactor responsive high proliferative potential-colony forming cells [HPP-CFCs]). Additionally, we confirm the ability of transduced progenitor cells to engraft into the normal, nonmyeloablated host.
MATERIALS AND METHODS
Bone marrow harvest.Bone marrow was harvested from the tibia and femur of male BALB/c mice (18-22 g, Charles River, Wilmington, MA) as previously described.19 Donors in the first cohort received 0.5 μg of recombinant murine stem cell factor (SCF; Genetics Institute, Cambridge, MA) in 100 μL of Dulbecco's Phosphate-Buffered Saline (DPBS) by intraperitoneal (IP) injection 24 hours before collection of marrow. Cells were dispersed by pipetting with a 10-mL pipette, nucleated-cell counts obtained in a 19:1 dilution with crystal violet, centrifuged at 1,200 rpm for 10 minutes, and resuspended in DPBS (GIBCO, Grand Island, NY) to the appropriate concentration.
MDR18.1 and N2-5 producer cell line.The cell line MDR18.1 produces the human MDR1 cDNA incorporated into an ecotropic retroviral vector driven by the Harvey murine sarcoma virus long terminal repeats,24 27 and the N2-5 cell line produces vector that contains the Neomycin resistance gene (both obtained from David Bodine, National Institutes of Health, Bethesda, MD). The MDR18.1 and N2-5 cell lines were maintained at 37°C 5% CO2 in growth medium: Dulbecco's modified Eagle's medium (DMEM) low glucose (GIBCO), 4 mmol/L glutamine (GIBCO), penicillin-streptomycin (GIBCO), and 15% heat-inactivated newborn (less than 10 days) bovine serum (GIBCO).
Incubations (prestimulation and transduction).Bone marrow cells were cultured at 1 × 106/mL in infection medium: DMEM low glucose (GIBCO), 4 mmol/L glutamine (GIBCO), penicillin-streptomycin (GIBCO), and 15% heat-inactivated fetal bovine serum (Hyclone, Logan, UT) in Corning (Corning Inc, Corning, NY) suspension dishes (#25070-100). The 48-hour prestimulation was done in medium alone or in the presence of a cytokine cocktail, which contained 50 U/mL of interleukin-3 (IL-3), 50 U/mL IL-6, 50 ng/mL recombinant human IL-11 (Genetics Institute), and 50 ng/mL recombinant murine SCF. Cells were harvested and resuspended in a small volume of DPBS. Cells were cocultured with retroviral producer cells for 24 hours in the same medium and cytokine mixture as the prestimulation at the same concentration (1 × 106/mL). Polybrene (Hexadimethrine Bromide from Sigma, St Louis, MO) was added to the retroviral cocultures at a final concentration of 6 μg/mL.
HPP-CFC assay.Bone marrow cells (control or transduced) were plated in the HPP-CFC assay as previously described.28 29 Briefly, for each 35-mm dish 25,000 or 50,000 cells are plated into the top agar layer (0.5 mL) over a bottom feeder layer of agar, medium, and cytokines (1.35 mL). The cytokine mixture used for this in vitro hematopoietic progenitor colony assay is given as the concentration in the underlayer, and the final concentration for the entire dish is listed in brackets after. The 6-factor plates contained 52,000 U [38,000 U] human colony-stimulating factor-1 (CSF-1; Cetus, Emeryville, CA), 26 ng [19 ng] murine granulocyte-macrophage colony-stimulating factor (GM-CSF; Amgen, Thousand Oaks, CA), 52 ng [38 ng] human granulocyte colony-stimulating factor (G-CSF; Amgen), 1,040 U [760 U] murine IL-3 (Collaborative, Bedford, MA), 2,600 U [1,900 U] human IL-1a (HoffmannLaRoche, Nutley, NJ), and 1.04 μg [0.76 μg] murine SCF (R & D Systems, Minneapolis, MN); for the 7-Factor plates, 52 ng [38 ng] of bovine basic FGF (Collaborative) was also included. The high–CSF-1 plates each received only 200,000 U [146,000 U] of CSF-1. Plates were incubated at 37°C, high humidity, 5% oxygen, and 10% CO2 for 7 to 14 days. The HPP-CFC was defined as a tight ball of cells at 14 days of greater than 0.5 mm in diameter or a dense colony greater than 1.0 mm in diameter.
Colony reverse transcriptase-polymerase chain reaction (RT-PCR) assay.Individual, nonoverlapping HPP-CFC colonies were picked from the agar using a solution of sterile water with 0.2 mg/mL brewers yeast tRNA (Boehringer Mannheim, Mannheim, Germany). Colony RNA was prepared by a variation of the method of Witesell and Schook.30 Briefly, 200 μL of a guanidine isothiocyanate (GSCN) solution (4 mol/L GSCN, 50 mmol/L Tris-HCl pH 7.5, 25 mmol/L EDTA [GIBCO Catalog #5577UA] with 0.75% 2-mercaptoethanol and 0.5% NP-40) was added to each sample, vortexed for 10 seconds, and 25 μL of 2 mol/L sodium acetate (pH 4.5) was added. Then each tube received 350 μL of 50% phenol/49% chloroform/1% isoamyl alcohol. Samples were vortexed for 10 seconds, ice chilled for 15 minutes, and microcentrifuged at 14,000 rpm for 2 minutes. The RNA was precipitated with isopropanol for 4 hours at −20°C, microcentrifuged at 14,000 rpm for 15 minutes, drained, and dried in a Speed Vac Evaporator (Savant Instruments Inc, Holbrook, NY). Each pellet was resuspended in 200 μL of TE-NaCl (0.2 mol/L NaCl, 10 mmol/L Tris, 1 mmol/L EDTA, pH 7.2) then precipitated in 800 μL of ethanol for at least 4 hours at −20°C. RNA samples were pelleted at 14,000 rpm for 15 minutes, dried, and then resuspended in 44 μL of sterile pure water (GIBCO) for use.
The reverse transcriptase (RT) reaction was performed using 10 μL of the colony RNA added to 14.5 μL of PCR-grade water, 3.5 μL of 10 U/μL RNasin (Promega, Madison, WI), and 2 μL of 10 nmol/L random primers (GIBCO). After heating at 70°C for 5 minutes, samples were ice chilled and to each was added 3.5 μL of RNasin at 10 U/μL, 13 μL of 10X SSRT buffer (GIBCO), 17.5 μL of 2 mmol/L dNTPs, and 1 μL of SSRTII RT (GIBCO). Samples were heated to 42°C for 60 minutes, heated to 70°C for 5 minutes, and then either stored at −20°C or processed immediately in the PCR reaction.
The PCR amplification reaction of the cDNA was performed on one third of each RT reaction (20 μL), allowing for sufficient template to run duplicate samples and a control, usually a ”no-primer“ reaction, for each colony. The modified “hot-start” PCR reactions contained template and primer in the bottom layer, a layer of paraffin wax (Wax Gem 100, Perkin Elmer-Cetus, Norwalk, CT), and the reaction mix containing buffer, nucleotides, and enzyme on the top. Primers were used at a final concentration of 1 μmol/L and final reaction conditions were 50 mmol/L KCl; 10 mmol/L Tris-HCl pH 8.3; 3.25 mmol/L MgCl2; 0.2 mmol/L each dATP, dCTP, dGTP, and dTTP; AmpliTAQ TAQ polymerase (Perkin Elmer), and 4 μCi 32 PdCTP (NEG-O37Z, NEN; DuPont, Wilmington, DE). PCR used 35 cycles of 94°C for 1 minute, 55°C for 1 minute, and 72°C for 2 minutes. Primers used for PCR24: MDR (upstream) 5′-GCC CAC ATC ATC ATG ATC-3′; MDR (downstream) 5′-GTC TCC TAC TTT AGT GCT-3′. For analysis, samples were resolved on a 5% acrylamide gel and bands were measured by ultraviolet (UV) shadow, then gels were dried onto 3MM Whatman paper (Whatman International, Kent, UK) and exposed to radiography film.
Transplant into nonmyeloablated hosts.Transduced male BALB/c bone marrow cells were harvested after the retroviral incubation and washed in Hank's buffered saline solution (HBSS; GIBCO) three times, then resuspended into DPBS for injection via tail vein into normal female BALB/c mice; injection volumes ranged from 0.3 mL to 0.62 mL/mouse. The animals that completed the study were (cell dose per day on consecutive days is listed after each): TX1 (20 × 106, 20 × 106), TX2 (12 × 106, 12 × 106, 12 × 106, 12 × 106, 12 × 106), TX5 (10 × 106, 20 × 106, 14 × 106), TX6 (20 × 106, 15 × 106), and TX40 and TX60 (DPBS each day for 5 days, same volume as cell infusions). Mice were given 10 mg/kg TAXOL (IP) at three times over a 14-month period. TAXOL was supplied by Bristol Myers/Squibb (Princeton, NJ), and delivered at 5 and a half months, 9 and a half months, and 13 months posttransplant. Animals were sacrificed and analyzed 2 weeks after the last TAXOL injection.
Analyses of transplant recipients.The DNA from tissues and organs was analyzed by Southern blot for the presence of Y chromosome specific sequences as previously described.19 Briefly, DNA was digested with Dra I restriction endonuclease, resolved on 0.8% agarose gel, transferred in 0.4Normal NaOH to ZetaProbe (Bio-Rad, Richmond, VA), and probed for male specific sequence with the PY2 plasmid31; gel loading was corrected using an IL-3 cDNA probe that recognizes an autosomal single copy gene.32 Signal from Southern blots was quantified using Radioanalytic Phosphorimaging (Molecular Dynamics, Sunnyvale, CA). Initially, the DNAs were also analyzed for the presence of the MDR1-containing proviral DNA by Southern blot; however, the analysis was extended to include a more sensitive PCR assay using MDR1-specific primers (listed previously) and an internal reaction and copy number control for murine β-globin [bg1 (upstream) 5′-GAA GTT GGG TGC TTG GAG AC-3′; bg2 (downstream) 5′-GAG ACT GCT CCC TAG AAT CG-3′]. Whole cell preps from bone marrow and peripheral blood were also assayed by fluorescent in situ hybridization (FISH) using a digoxygenin-labeled murine Y chromosome painting probe developed by U. Weier33 to selectively stain male bone marrow cells in the transplant recipients.20 The presence of mRNA from the transferred gene was evaluated using the same RT-PCR assay as described for colony RNAs and 3 to 5 μg of total cellular RNA as template.
RESULTS
To evaluate the percent of hematopoietic progenitor cells that were transduced effectively, we assayed for the expression of the vector-transferred gene mRNA by RT-PCR from individual HPP-CFC colonies. The HPP-CFC is an in vitro surrogate representing a multilineage hematopoietic progenitor/stem cell population that correlates with marrow renewal.28,29 The HPP-CFC is the predominant cell, approaching 100%, in certain ”purified“ stem cell populations including the Hoechstlow Rhodaminelow lineage negative cells34 indicating that it is an in vitro surrogate for an early hematopoietic progenitor cell population. We have chosen to use the HPP-CFC assay, which represents an early progenitor and which lends itself easily to colony RT-PCR analysis.
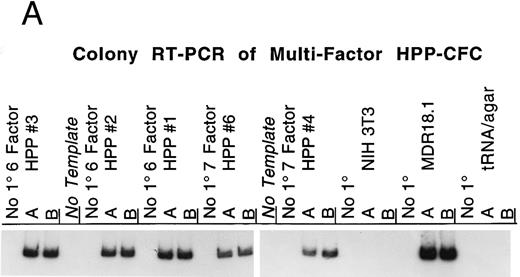
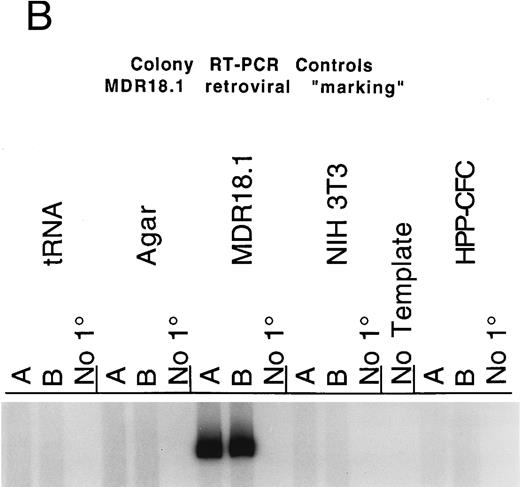
The HPP-CFC plating assay routinely includes six cytokines (SCF, CSF-1, IL-1α , IL-3, G-CSF, and GM-CSF ), and often basic FGF is added as well. Transduction of these progenitors was determined by RT-PCR of individual colonies to detect RNA from the transferred human MDR1 gene. Figure 1A shows examples of RT-PCR using MDR1 primers and RNA from HPP-CFC colonies. The cDNA from each colony RT reaction is split into thirds, allowing for one control reaction and duplicate analytical reactions. The colonies selected for analysis had opaque centers 1.0 to 2.0 mm in diameter. Additionally, a group of differentiated macrophage colonies, grown in serum plus CSF-1, was screened and 50% showed expression of the MDR1 mRNA (Figs 2 and 3). The primers used are located upstream of a cryptic splice site in the MDR1 cDNA, thereby avoiding detection problems associated with the shortened mRNA that is sometimes generated (D. Bodine, personal communication); in our hands this is not a problem as we get mRNA and protein expression from the transgene. The routine positive and negative controls that are needed for this assay are shown in Fig 1B. The colony RT-PCR assay reproducibly detects the RNA transcript from the human MDR1 cDNA. Controls show no signal from the possible sources of background, eg, mouse RNAs, tRNA carrier, and agar from between colonies.
(A) RT-PCR detection of MDR1 expression by multifactor HPP-CFC. Single murine HPP-CFC colonies were picked and assayed for expression of the transferred human MDR1 gene using the RT-PCR assay as described in Materials and Methods. The first set of three lanes is from a 6-factor HPP-CFC (HPP#3) transduced with the MDR1 vector. This colony, as well as HPP#2, HPP#1, HPP#6, and HPP#4, all contain the RT-PCR fragment corresponding to the mRNA from the transferred human MDR1 vector sequence. Each lane represents RNA equivalent to of a colony. Samples are run in duplicate with a no-primer (no 1°) control. No-template negative control reactions are interspersed among the assay samples. The last sets are NIH3T3 RNA (negative control, murine cell line), MDR18.1 RNA (positive control, MDR1 retroviral producer line), and the tRNA/agar control for background (negative control for nonspecific signals and the carrier in the RNA isolation protocol). (B) Colony RT-PCR assay controls. This panel shows the results of the control templates routinely run during the RT-PCR assay for MDR1 expression. Duplicate control templates are amplified concurrently with test samples using the same reagents and conditions; a no-primer (no 1°) control is also included for each. Control templates (from left to right) are (1) tRNA carrier, which is present in the RNA extractions: This gives no RT-PCR background when SSRTII is used for reverse transcription; (2) agar from between colonies: Neither the agar nor any extraneous material from the transduction or plating generates RT-PCR background; (3) MDR18.1 RNA from the producer cell line: 1 μg of total cellular RNA in the RT step is included as a positive control; (4) NIH3T3 cell line RNA (or murine bone marrow RNA): 1 μg of total cellular RNA in the RT step is included as a negative control (this is done to detect any cross reaction with any of the MDR isoforms present in mice); (5) No-template reaction: This is a standard PCR control for each set of primers used; and (6) a nontransduced multifactor murine HPP-CFC colony: This is processed identically to the experimental samples so that each lane represents of a colony.
(A) RT-PCR detection of MDR1 expression by multifactor HPP-CFC. Single murine HPP-CFC colonies were picked and assayed for expression of the transferred human MDR1 gene using the RT-PCR assay as described in Materials and Methods. The first set of three lanes is from a 6-factor HPP-CFC (HPP#3) transduced with the MDR1 vector. This colony, as well as HPP#2, HPP#1, HPP#6, and HPP#4, all contain the RT-PCR fragment corresponding to the mRNA from the transferred human MDR1 vector sequence. Each lane represents RNA equivalent to of a colony. Samples are run in duplicate with a no-primer (no 1°) control. No-template negative control reactions are interspersed among the assay samples. The last sets are NIH3T3 RNA (negative control, murine cell line), MDR18.1 RNA (positive control, MDR1 retroviral producer line), and the tRNA/agar control for background (negative control for nonspecific signals and the carrier in the RNA isolation protocol). (B) Colony RT-PCR assay controls. This panel shows the results of the control templates routinely run during the RT-PCR assay for MDR1 expression. Duplicate control templates are amplified concurrently with test samples using the same reagents and conditions; a no-primer (no 1°) control is also included for each. Control templates (from left to right) are (1) tRNA carrier, which is present in the RNA extractions: This gives no RT-PCR background when SSRTII is used for reverse transcription; (2) agar from between colonies: Neither the agar nor any extraneous material from the transduction or plating generates RT-PCR background; (3) MDR18.1 RNA from the producer cell line: 1 μg of total cellular RNA in the RT step is included as a positive control; (4) NIH3T3 cell line RNA (or murine bone marrow RNA): 1 μg of total cellular RNA in the RT step is included as a negative control (this is done to detect any cross reaction with any of the MDR isoforms present in mice); (5) No-template reaction: This is a standard PCR control for each set of primers used; and (6) a nontransduced multifactor murine HPP-CFC colony: This is processed identically to the experimental samples so that each lane represents of a colony.
Hematopoietic progenitor expression of MDR1 in vitro. Hematopoietic progenitor colonies, HPP-CFC, from bone marrow transduced with the MDR18.1 retroviral vector were analyzed for expression of the MDR1 RNA using the RT-PCR assay. From a group of 99 colonies, 48 were generated by bone marrow transduced in the presence of cytokines (IL-3, IL-6, IL-11, and SCF ), and 51 were generated by bone marrow transduced in the absence of cytokines. As shown in the graph on the left, both groups were efficiently transduced. The transduction in the presence of cytokines (+) results in 83% of colonies positive for MDR1, and transduction in the absence of added cytokines (−) results in 92% of the colonies scoring positive. After transduction (+ or − cytokines), bone marrow cells were plated in either the multifactor HPP-CFC assay or in CSF-1 alone. Of the 99 colonies analyzed, 81 were generated from the HPP-CFC assay and 18 were generated from the CSF-1 platings; these are compared in the graph on the right. Colonies from the multifactor HPP-CFC assay are multilineage and highly proliferative (<1.0 mm in diameter); 96% of these were positive for expression of MDR1 RNA. The colonies grown in CSF-1 alone were predominantly macrophages, and were seldom greater than 0.5 mm in diameter; 50% of these were positive for expression of the MDR1 RNA. When the results of both transduction methods and both plating assays are combined, the overall transduction rate into hematopoietic progenitors is 88%.
Hematopoietic progenitor expression of MDR1 in vitro. Hematopoietic progenitor colonies, HPP-CFC, from bone marrow transduced with the MDR18.1 retroviral vector were analyzed for expression of the MDR1 RNA using the RT-PCR assay. From a group of 99 colonies, 48 were generated by bone marrow transduced in the presence of cytokines (IL-3, IL-6, IL-11, and SCF ), and 51 were generated by bone marrow transduced in the absence of cytokines. As shown in the graph on the left, both groups were efficiently transduced. The transduction in the presence of cytokines (+) results in 83% of colonies positive for MDR1, and transduction in the absence of added cytokines (−) results in 92% of the colonies scoring positive. After transduction (+ or − cytokines), bone marrow cells were plated in either the multifactor HPP-CFC assay or in CSF-1 alone. Of the 99 colonies analyzed, 81 were generated from the HPP-CFC assay and 18 were generated from the CSF-1 platings; these are compared in the graph on the right. Colonies from the multifactor HPP-CFC assay are multilineage and highly proliferative (<1.0 mm in diameter); 96% of these were positive for expression of MDR1 RNA. The colonies grown in CSF-1 alone were predominantly macrophages, and were seldom greater than 0.5 mm in diameter; 50% of these were positive for expression of the MDR1 RNA. When the results of both transduction methods and both plating assays are combined, the overall transduction rate into hematopoietic progenitors is 88%.
MDR1-positive colonies from different transduction medium and plating conditions. The transduction results from a group of 99 hematopoietic colonies, previously described in Fig 2, is shown here in grid format. The colonies are divided into transduction conditions, four cytokines or in medium alone, along the vertical axis. Cells are then further divided, along the horizontal axis, into the colony assay conditions in which they were grown, HPP-CFC (6 or 7 factor) and high–CSF-1. The number of colonies in each group appears in parentheses. The percent positive is listed in bold in each section. This illustrates the efficient transfer of the vector into the HPP-CFC regardless of transduction conditions or plating assay; colonies are 83% to 97% positive. Interestingly, both transduction conditions resulted in lower transfer into the macrophage colonies; 45% were positive from the four cytokine transductions and 57% were positive from the transduction in medium alone.
MDR1-positive colonies from different transduction medium and plating conditions. The transduction results from a group of 99 hematopoietic colonies, previously described in Fig 2, is shown here in grid format. The colonies are divided into transduction conditions, four cytokines or in medium alone, along the vertical axis. Cells are then further divided, along the horizontal axis, into the colony assay conditions in which they were grown, HPP-CFC (6 or 7 factor) and high–CSF-1. The number of colonies in each group appears in parentheses. The percent positive is listed in bold in each section. This illustrates the efficient transfer of the vector into the HPP-CFC regardless of transduction conditions or plating assay; colonies are 83% to 97% positive. Interestingly, both transduction conditions resulted in lower transfer into the macrophage colonies; 45% were positive from the four cytokine transductions and 57% were positive from the transduction in medium alone.
We analyzed 99 bone marrow HPP-CFC colonies from cells transduced using the MDR18.1 retroviral producer line in the presence of the four cytokine cocktail (IL-3, IL-6, IL-11, and SCF ) or medium alone. Cells were harvested and plated in either 6- or 7-factor HPP-CFC agar or in agar with high–CSF-1 alone. From the cells cultured in the presence of the 4-cytokine cocktail, we selected 48 colonies. From the cells cultured in the absence of the additional cytokines, we selected 51 colonies. Cells were plated in either 6-factor (n = 53 colonies selected), 7-factor (n = 64 colonies selected), or high–CSF-1 (n = 81 colonies selected) agar bilayers for 7 or 14 days. All colonies combined (n = 99) resulted in an 88% transduction rate as determined by 87 of 99 colonies scoring positive for mRNA transcript from the MDR1 transgene. Figure 2 shows the colonies by transduction group (in the presence or absence of added cytokines) and by plating groups. The transduction conditions did not result in any significant difference in expression of the MDR1 RNA by HPP-CFC colonies. Interestingly, the more mature macrophage colonies on the high–CSF-1 plates were transduced at a lower efficiency (50%) as compared with the other transduction rates. Figure 3 summarizes the results of each combination of transduction conditions and plating type variables in a matrix.
Long-term engraftment of transduced progenitor cells into normal hosts.For evaluation of engraftment of transduced cells into the normal marrow microenvironment, the male bone marrow cells were collected after transduction and injected into normal nonmyeloablated female BALB/c hosts in a solution of DPBS. Mice were followed for 14 months posttransplant by PCR of peripheral blood before and after each of the three TAXOL (10 mg/kg IP) injections, using MDR-specific primers. At each time point, all PCR analyses of peripheral blood samples were negative for MDR1 proviral sequences. At 14 months, 2 weeks after the third Taxol injection, host tissues were harvested and analyzed for both Y-specific sequence and presence of the MDR provirus.
Analysis of bone marrow by FISH, using a male-specific probe showed donor (ie, male) engraftment of the ex vivo manipulated marrow. The engraftment as determined by Y-chromosome FISH shows engraftment levels between 8.9% and 13.4%. Slides were visually scored by three different investigators and then averaged. Southern blot with phosphorimage image analysis, resulted in calculated engraftment values that were close to, but not identical to those obtained by FISH: TX1 9.1%, TX2 7.0%, TX5 6.5%, TX6 7.1%, and TX40 and TX60 0%. Results are summarized in Table 1. Engraftment in host splenic tissues from the same mice when determined by Southern blot was lower than in bone marrow: TX1 4.2%, TX2 0.6%, TX5 0.7%, and TX6 2.9%.
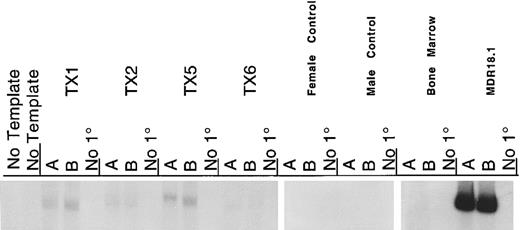
PCR amplification was used to detect the presence of MDR1 proviral DNA; this showed no detectable MDR1 DNA in the peripheral blood taken before and after each Taxol challenge. Additionally, there was no MDR1 DNA detected in the bone marrow or spleen tissues harvested at 14 months (Fig 4). Using RT-PCR to detect expression of mRNA we did detect the MDR1 mRNA in bone marrow, although at very low levels as shown in Fig 5. From a dilution curve prepared with MDR-transduced cells mixed with noninfected cells, we estimate the number of cells producing MDR transcript to be between 1 in 10,000 and 1 in 100,000. We conclude that our low expression of the transgene was not a result of gene “shut-off” because the proviral DNA was not detected. Rather the transgene is present and expressed in only a small number of engrafted donor cells. Although this demonstration is not stringently quantitative, it is an exciting confirmation that successfully transduced cells were engrafted in the normal host.
Analysis of host bone marrow DNAs for vector DNA. Bone marrow DNA from transplant recipients and controls was analyzed by PCR for the presence of the MDR18.1 vector DNA. For each reaction, 250 ng of template DNA was amplified with the human MDR1 primers and the murine β-globin primers using the same PCR reaction conditions as the MDR1 colony RT-PCR assay. The murine β-globin serves as an internal standard for copy number and quality of template. The MDR1 fragment is the upper band, and the β-globin is 100 bases shorter and runs below the MDR1. This assay is capable of detecting 1 copy of the transgene per haploid genome in <1% of the cells as determined by a standard dilution curve. None of the transplant recipients showed detectable amounts of MDR1 DNA using this assay. The templates assayed are (from left to right) two no-template negative control reactions; NIH3T3 (murine cell line; negative control for human MDR1 gene); MDR18.1 (the retroviral producing murine cell line; positive control, producer line contains the proviral genome); male murine spleen DNA (negative control); TX40 and TX60 bone marrow DNAs (negative control; these mice received no MDR-transduced cells, only phosphate-buffered saline); and TX1, TX2, TX5, TX6 bone marrow DNAs (transplant recipients).
Analysis of host bone marrow DNAs for vector DNA. Bone marrow DNA from transplant recipients and controls was analyzed by PCR for the presence of the MDR18.1 vector DNA. For each reaction, 250 ng of template DNA was amplified with the human MDR1 primers and the murine β-globin primers using the same PCR reaction conditions as the MDR1 colony RT-PCR assay. The murine β-globin serves as an internal standard for copy number and quality of template. The MDR1 fragment is the upper band, and the β-globin is 100 bases shorter and runs below the MDR1. This assay is capable of detecting 1 copy of the transgene per haploid genome in <1% of the cells as determined by a standard dilution curve. None of the transplant recipients showed detectable amounts of MDR1 DNA using this assay. The templates assayed are (from left to right) two no-template negative control reactions; NIH3T3 (murine cell line; negative control for human MDR1 gene); MDR18.1 (the retroviral producing murine cell line; positive control, producer line contains the proviral genome); male murine spleen DNA (negative control); TX40 and TX60 bone marrow DNAs (negative control; these mice received no MDR-transduced cells, only phosphate-buffered saline); and TX1, TX2, TX5, TX6 bone marrow DNAs (transplant recipients).
RT-PCR detection of MDR1 in transplant recipient bone marrow. RNA was harvested from bone marrows of the transplanted mice at 14 months. Each RT reaction with 3 μg of total cellular RNA was split into three PCR reactions, these included two parallel analytical reactions and a no primer (no 1°) negative control. PCR conditions were as described for colony RT-PCR. As shown in this autoradiogram, there is low, but detectable, MDR transcript in each of the four transplant recipient mice. Controls include female and male murine spleen RNA from nontransplanted mice, murine bone marrow RNA from nontransplanted mice as negative controls, and RNA from the MDR18.1 producer line as positive control.
RT-PCR detection of MDR1 in transplant recipient bone marrow. RNA was harvested from bone marrows of the transplanted mice at 14 months. Each RT reaction with 3 μg of total cellular RNA was split into three PCR reactions, these included two parallel analytical reactions and a no primer (no 1°) negative control. PCR conditions were as described for colony RT-PCR. As shown in this autoradiogram, there is low, but detectable, MDR transcript in each of the four transplant recipient mice. Controls include female and male murine spleen RNA from nontransplanted mice, murine bone marrow RNA from nontransplanted mice as negative controls, and RNA from the MDR18.1 producer line as positive control.
DISCUSSION
These data show that the majority of murine hematopoietic progenitor cells are transduced with the retroviral vector-delivered human MDR1 gene. In excess of 88% of HPP-CFC produce mRNA for the transgene. This colony assay reflects a primitive pluripotential population of hematopoietic cells, which correlates with host marrow repopulation,34 and we conclude that standard transduction procedures, which use ex vivo culture and cytokines to facilitate retroviral integration, are successful in transfecting this primitive cell population.
While demonstration of effective gene transfer into the target cell population is critical to applying this model, it is even more important to show that the cells responsible for long-term in vivo engraftment can be effectively transduced. These data show that transduced hematopoietic cells expressing the human MDR1 gene are capable of long-term engraftment in a nonablated host marrow. This has exciting applications for gene therapy of genetic disorders and diseases such as HIV that do not necessitate destruction of the host bone marrow.
The nonablated bone marrow microenvironment may constitute a qualitatively different engraftment site than the ”empty“ or altered marrow resulting from chemotherapy or radiation. In an ablated marrow, repopulation is essentially oligoclonal, the result of expansion of a small subpopulation of engrafted progenitors. The use of nonmyeloablated hosts was initally reported by Brecher et al22 and has been subsequently used to characterize the functional status of engrafting hematopoietic stem cells.19-21,35 Using this model, donor progenitor cell engraftment has been reproducibly shown to be truly competitive in this environment. Donor cells labeled with PKH20 show superior engraftment in the normal host marrow, indicating that the nonmyeloablated marrow microenvironment may even have advantages over the ablated host marrow.36
The donor cell engraftment levels are in an acceptable range for use in gene transfer procedures in nonablated hosts (8% to 10%). The number of progenitor cells that are transduced with the MDR1 gene is also within a range that would facilitate efficient gene transfer, as shown by the high rate of HPP-CFC scoring positive for MDR1 mRNA. However, the number of effectively transduced progenitor cells that engrafted is quite low. These data suggest that the majority of progenitor cells that integrated the MDR1 gene failed to engraft and that, of the cells that did engraft, only a small fraction had integrated the transgene. Thus, we propose that exposing the donor bone marrow to the retroviral transduction procedure impairs engraftment of transduced hematopoietic progenitors in the nonablated host.
To facilitate retroviral gene insertion, hematopoietic target cells are frequently stimulated to proliferate in vitro with cytokines. Cytokine stimulation induces the cell cycle transit necessary for successful proviral integration into the genomic DNA of the most primitive cells that are quiescent, or in G0 phase of the cell cycle.37-43 This procedure is intended to induce proliferation of early hematopoietic progenitor cells without inducing significant differentiation. The progenitor population assayed by HPP-CFC colony formation is an early multilineage hematopoietic cell that is quiescent and is stimulated to proliferate in vitro when plated in the agar medium and cytokines. Recent data from our group have shown that culture in cytokines results in the entry into active cell cycle transit by these HPP-CFC forming cells.35 This entry into cell cycle by bone marrow progenitors results in impaired long-term in vivo engraftment in nonmyeloablated hosts regardless of whether cell-cycle progression is induced by ex vivo culture in cytokines or by pretreatment of donors with drugs such as 5-Fluorouracil.20,35 A competitive engraftment model using irradiated murine hosts resulted in loss of repopulation potential of the cycling cells from cytokine-stimulated cultures after a few hours,44 thus showing defective long-term engraftment in the ablated marrow as well. Taken together, these data show a strong correlation between active cell-cycle progression and loss of engraftment potential.
The engraftment defect correlated with the transduction procedures is not limited to murine bone marrow transduced with the human MDR1 vector, MDR18.1. In additional studies (data not shown), BALB/c mice that received bone marrow transduced with the neomycin resistance gene, retroviral vector N2-5, also showed no detectable transfer of the transgene by standard Southern blot.
Engraftment of murine bone marrow progenitors transduced with a human MDR1 may also be influenced by host immunity. Although cell cycle considerations are compelling, it is important to note that in a nonmyeloablated host, the immune system may affect the selection of engrafting cells. In additional studies, sera from MDR1 transplant recipients showed no specific detectable response to cells expressing human MDR1 on their surface when assayed by fluorescence-activated cell sorting (data not shown). Additionally, the response of splenic T-cells from the MDR1 transplant recipients to stimulation by a crude extract of MDR18.1 cells did not exceed the standard stimulation index (proliferation score divided by control score) significance value of 2.000 (data not shown). This lack of detectable immune system recognition or response to human MDR1 indicates that failure of MDR1 transduced marrow to engraft in the nonablated host is probably not related to host immunity.
Cytokine-stimulated proliferation is implicated as the mechanism that induces a functional change in the progenitor cells that is related in some way to their cell-cycle status. This cell cycle related engraftment defect has been shown in nonablated hosts, as well as irradiated hosts.20,35,44 Interestingly, in these studies that used the cytokine incubation to facilitate retroviral integration, we achieved superior levels of engraftment to those reported previously by Peters et al.35,44 The retroviral transduction begins with the same cytokine incubation but is then followed by a 24-hour coculture with the retroviral producer cell line that is of murine fibroblast origin. We speculate that this might offer the progenitor cells a needed cellular factor (conditioned medium) or simply be the result of cell-cell contact. It is an exciting possibility for providing an atmosphere that either maintains or even reestablishes engrafting potential. To this end, many gene transfer protocols are investigating the use of autologous stromal components during ex vivo gene transfer. Transfer into a nonmyeloablated canine host was achieved with transduced long-term bone marrow cultures; the transduction of cells cultured on autologous stroma resulted in retention of engraftment ability.9 This manner of transduction, along with other approaches, may be critical to maximizing the number of long-term progenitors that retain engraftment potential while being effectively transduced.
The cellular changes induced by proliferation are evidenced by defective engraftment, but may also reflect differentiation as well as proliferation. The two processes are intricately linked during development and maturation of many cell types including hematopoietic cells. Although a portion of cells may have been induced to differentiate, the engraftment of donor cells, which is detected at 14 months posttransplant, shows that a subset of long-term repopulating cells was contributing to engraftment of the transplanted marrow. Nonablated mice carrying the mutation W/Wv also show low engraftment of transduced donor cells, particularly those carrying the NeoR transgene.10 Although it was postulated that the low engraftment was caused by nonablation and no preselection in G418, we speculate that the in vitro transduction culture was responsible for the engraftment defect also evident in the defective microenvironment of the W/Wv hosts. This in combination with the other data from this group20,35 44 indicates that the majority of engrafting cells may be derived from a population of stem cells that were not actively cycling during the transduction incubations.
These data also show equivalent high transduction rates into marrow progenitor cells when retroviral transduction is performed in the absence of cytokines. Proliferation is required for proviral integration, but it is very likely that retrovirus can enter the cytoplasm of a noncyling cell. The cell proliferation stimulated by the conditions of the ensuing HPP-CFC assay may allow integration of the provirus in vitro, thus accounting for the high transduction efficiency. Similarly, the engrafted progenitors that did express the transgene may not have been proliferating during transduction, but the vector was integrated at a later time in vivo, or after transplant into the host. This is consistent with evidence from Visser et al36 who have shown that a majority of purified stem cells engrafted in vivo proliferate within 17 hours of engraftment. Additionally, human clinical trials using short viral incubation times without cytokines have achieved expression of the transgene in children.45 Although this included introduction into an ablated marrow, it does suggest that in vitro incubation with cytokines before stem cell transplant may be unnecessary.
This report establishes the transfer of the human MDR1 gene into the nonmyeloablated murine host. Whereas the demonstration of gene transfer in this model has exciting implications for gene therapy, the importance of maintaining engraftment potential is strongly emphasized. These studies show that stimulation of cell proliferation as a means to facilitate retroviral vector integration results in an engraftment defect that corresponds to the induction of cell cycle transit. We conclude that this gene-delivery system is capable of transducing hematopoietic stem/progenitor cells, but that a relatively small fraction of these cells are capable of engrafting and contributing to the hematopoietic system of a nonmyeloablated host under the conditions of these experiments. Maintaining the engraftment potential, either by culture in a suitable microenvironment or by reversing the loss of engraftment potential, will be an important step in applications of hematopoietic stem cell transplantation and gene transfer.
ACKNOWLEDGMENT
The authors thank David Bodine for the cell lines, MDR18.1 and N2-5, and many helpful discussions. Cheryl Y. Tiarks, Judith Reilly, Gerri Riel, and Bernarda Fraioli provided excellent technical support. A special thanks is given to Philip A. Lowry, Walter Demkowicz, Maria L. Zapp, and Cathleen L. Cooper for comments and consultation.
Supported by research grants from the National Institutes of Health (#PO1.CA27466), the American Cancer Society (#DHP-96), and the University of Massachusetts Medical Center.
Address reprint requests to Ellen L.W. Kittler, PhD, University of Massachusetts Cancer Center, Two Biotech, 373 Plantation Street, Suite 207, Worcester, MA 01605.