Abstract
Genes upregulated by p53 were screened using an erythroleukemic cell line (1-2-3) that expresses only the temperature-sensitive p53 by the mRNA differential display method. One of the upregulated genes was identified as the elongation factor-1α (EF-1α) gene, an essential component of the eukaryotic translation apparatus. Three p53-responsive elements were found in the mouse EF-1α gene and in the corresponding human, rat, and frog genes. These elements conferred the capacity for induction by p53. EF-1α is also a microtubule-severing protein. Upon the temperature-shift, the cells developed the morphology and the localization of α-tubulin similar to those of the cells treated with vincristine, a drug that affects microtubules. The microtubule-severing associated with upregulation of EF-1α by p53 may be a cause of the cell death.
THE TUMOR-SUPPRESSOR p53 gene, one of the most frequently mutated gene in human cancers,1 is a transcriptional factor that can bind to specific DNA sequences2 and increase gene expression from that element.3 Several p53-responsive genes, p21 (waf1/cip1/sdi1),4mdm2,5 mck,6 7 gadd45,8 cyclin G,9 bax,10 and insulin-like growth factor binding protein 311 have been identified. p21 acts as an inhibitor of cyclin-dependent kinases (CDKs), which are important regulators of the cell cycle.12-14 p21 has also been reported to bind to proliferating cell nuclear antigen (PCNA) and to inhibit DNA replication directly.15 The p21 gene is also identical to the Sdi1 gene that can induce cellular aging and inhibit DNA synthesis.16 Because mdm2 can bind to p53 and inactivate its activity as a transcription factor, it was proposed that mdm2 may act as a negative feedback regulator.5 Insulin-like growth factor binding protein 3 (IGF-BP3) is a negative regulator of the IGF-signaling. Upregulation of the IGF-BP3 gene by p53 may result in the growth-suppression of IGF-dependent cells.11 Wild-type p53 induces apoptosis of myeloid leukemic cells.17 Induction of apoptosis of thymocytes by ionizing radiation does not occur in p53-deficient mice.18,19 p53 also induces the apoptotic death of cells losing the normal retinoblastoma (RB) gene activity in the lens,20,21 retina,22 and brain.23 It has also been suggested that p53 might function to search for mutations in the RB gene and might induce apoptosis of cells containing mutations in this gene.24 bax can bind to bcl-2 that is an inhibitor of apoptosis and can accelerate apoptotic cell death.25 It was also reported that upregulation of the bax gene and downregulation of the bcl-2 gene by p53 are associated with apoptosis of a murine leukemic cell line M126 and that Fas, a transducer of a signal for apoptosis, is upregulated by p53 in non–small-cell lung adenocarcinoma and erythroleukemia.27
We established a murine erythroleukemic cell line (1-2-3) that expresses only a temperature-sensitive p53 protein (p53Val-135). The cell line had typical features of apoptosis when they were cultured at 32°C.28 Although the expression of the p21, gadd45, and cyclin G genes increased, no significant changes were detected in the expression of the mdm2, bcl-2, bax, fas, and fasl genes, suggesting that the existence of other genes associated with apoptosis.28 In this study, we screened other genes associated with cell death using the cell line using the mRNA differential display (DD) method29 and identified the elongation factor-1α (EF-1α) gene as an upregulated gene by p53. Although EF-1α is a polypeptide elongation factor that is an important component of the eukaryotic translation apparatus,30 EF-1α has also been identified as a microtubule-severing factor.31 We also discuss the association of cell death and microtubule-severing.
MATERIALS AND METHODS
Cell culture.The erythroleukemic cell line 1-2-3, which expressed only a temperature-sensitive mutant form of p53 (p53Val-135), was established previously.28 A second erythroleukemic cell line, 2-2, expressed another mutant p53 (not a temperature-sensitive protein).28 Tumor cells were cultured in RPMI 1640 (Flow Laboratories Inc, Irvine, UK) supplemented with 10% fetal bovine serum. For treatment with vincristine, cells were cultured in medium with 1 μmol/L vincristine (Sigma, St Louis, MO). For treatment with Taxol, cells were cultured in medium with 1 μmol/L Taxol (Sigma) for 1 hour and rinsed with fresh medium and then cultured without Taxol until they were tested for viability. For assays of viability, cells were stained with trypan blue and viable cells were counted.
DD.DD procedure was performed essentially as described by Liang and Pardee,29 using a Delta RNA Fingerprinting Kit (Clontech, Palo Alto, CA). Total RNA isolated from 1-2-3 cells cultured at 37°C for 24 hours and 32°C for 6 hours by the acid guanidium-thiocyanate-phenol-chloroform extraction method32 using Isogen (Nippongene, Toyama, Japan). Two micrograms of total RNA was reverse transcribed with 200 U of Moloney's murine leukemia virus reverse transcriptiase (Clontech) in the presence of 1 μmol/L of cDNA synthesis primer, 5 mmol/L dNTP mix for 1 hour at 42°C. cDNA samples were amplified by polymerase chain reaction (PCR) in the presence of 20 μmol/L of arbitrary primers (Clontech), 20 μmol/L of oligo (dT) primers (Clontech), 50 mmol/L of dNTP mix, 50 nm of [α-32P] dCTP (Amersham, Amersham, UK), and 2.5 U of Taq DNA polymerase (TOYOBO, Osaka, Japan). PCR primers used in this screening are listed below. The arbitrary primers were P1, ATTAACCCTCACTAAATGCTGGGGA; P2, ATTAACCCTCACTAAATGCTGGAGG; P3, ATTAACCCTCACTAAATGCTGGTGG; P4, ATTAACCCTCACTAAATGCTGGTAG; P5, ATTAACCCTCACTAAAGATCTGACTG; P6, ATTAACCCTCACTAAATGCTGGGTG; P7, ATTAACCCTCACTAAATGCTGTATG; P8, ATTAACCCTCACTAAATGGAGCTGG; P9, ATTAACCCTCACTAAATGTGGCAGG; and P10, ATTAACCCTCACTAAAGCACCGTCC. The Oligo (dT) primers were T1, CATTATGCTGAGTGATATCTTTTTTTTTAA; T2, CATTATGCTGAGTGATATCTTTTTTTTTAC; T3, CATTATGCTGAGTGATATCTTTTTTTTTAG; T4, CATTATGCTGAGTGATATCTTTTTTTTTCA; T5, CATTATGCTGAGTGATATCTTTTTTTTTCC; T6, CATTATGCTGAGTGATATCTTTTTTTTTCG; T7, CATTATGCTGAGTGATATCTTTTTTTTTGA; T8, CATTATGCTGAGTGATATCTTTTTTTTTGC; and T9, CATTATGCTGAGTGATATCTTTTTTTTTGG.
The conditions of PCR were 1 cycle of 94°C for 5 minutes, 40°C for 5 minutes, and 68°C for 5 minutes, followed by 2 cycles of 94°C for 5 minutes, 40°C for 2 minutes, and 68°C for 5 minutes, followed by 25 cycles of 94°C for 1 minute, 60°C for 1 minute, and 68°C for 2 minutes. Two microliters of denatured samples was fractionated in a 5% polyacrylamide gel at 70 W for 2.5 hours. Signals were detected by autoradiography. Differentially expressed bands were excised from the gel and boiled in TE buffer for 5 minutes. Solid debris was removed by centrifugation and the supernatant was used for reamplification by PCR with the same primers as DD. PCR was performed for 40 cycles (94°C for 1 minute, 60°C for 1 minute, and 72°C for 2 minutes).
Northern blot analysis.Twenty micrograms of total RNA was denatured in glyoxal and dimethyl sulfoxide, fractionated on 1% agarose gel, and transferred to a nylonmenbrane (Hybond N+; Amersham). After hybridization with 32P-labeled probes, signals were detected by autoradiography and intensities of bands were analyzed with a Bioimaging Analyzer (Bas2000; Fuji Film, Tokyo, Japan).
Cloning and DNA sequencing.Reamplified PCR fragments were cloned into the pCRII vector (Invitrogen, San Diego, CA). The sequence was determined by the dideoxy chain termination method33 with Sequencing PRO (TOYOBO). A homology search was performed with the FASTA program34 using GCG software (Wisconsin Sequence Analysis Package; Genetics Computer Group, Madison, WI).
Western blotting analysis.Cell lysates were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% polyacrylamide), transferred to a polyvinylidene difluoride (PVDF) membrane, and probed with a monoclonal antibody against EF-1α (UBI, Lake Placid, NY) using a Vectastain ABC kit (Vector, Burlingame, CA).
Luciferase assay.Reporter plasmids (pGVP-D9 and pGVP-E3E4) were constructed by the insertion of the DNA fragments D9 and E3E4 into the upstream of the luciferase gene of pGVP (Toyo Ink, same as pGL3-promoter; Promega, Madison, WI). 1-2-3 cells were transfected with reporter plasmids and cultured at 37°C or 32°C for 2 days. Luciferase activity was analyzed with the Dual-Luciferase Reporter Assay System (Promega). Saos-2 cells were transfected with reporter plasmids plus the p53-expression vector (pact-p53) or the control vector (pact1) and cultured for 2 days. Then luciferase activity was analyzed as mentioned above.
Staining of cells.For Giemsa staining, cultured cells were attached to glass slides, fixed with May-Grünwald's solution (Merck) and stained with Giemsa's solution (Merck, Whitehouse Station, NJ). For α-tubulin staining, cells were fixed and stained with monoclonal antibody against anti–α-tubulin (T-9026; Sigma) by the methods described previously.35 After staining with fluorescein isothiocyanate-conjugated second antibody, localization of α-tubulin was analyzed with fluorescence microscope.
RESULTS AND DISCUSSION
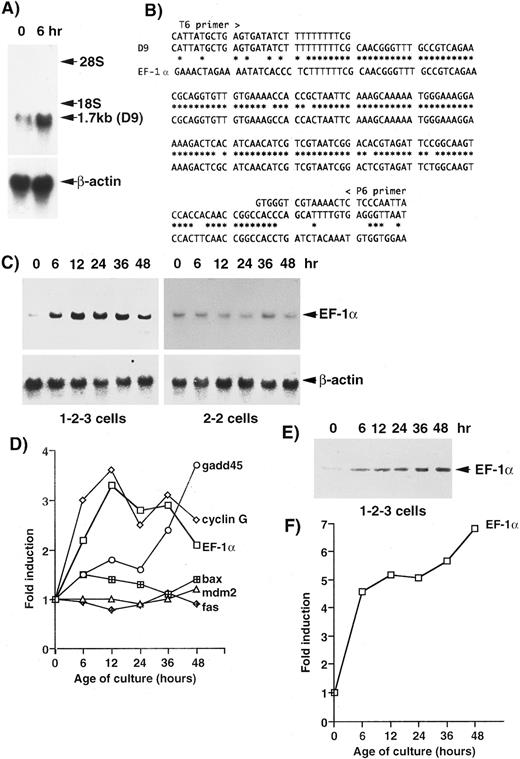
For screening other genes associated with cell death, cDNAs from 1-2-3 cells, which had been cultured at 37°C and 32°C for 6 hours, were amplified by PCR and analyzed by the mRNA DD method.29 One band, amplified by primers T6 and P6, was confirmed by Northern blotting analysis (Fig 1A) in this screening. The intensity of the band (1.7 kb) from cells cultured at 32°C for 6 hours was about twice of that from cells cultured at 37°C. The cDNA was reamplified by PCR and cloned into the pCRII vector. The sequencing of a clone (D9) that contained a 189-bp insert showed significant homology to the mouse EF-1α gene (Fig 1B). In 1-2-3 cells, the level of the expression of the EF-1α gene was about twice of that at 37°C after 6 hours and three times of that at 37°C (peak) after 12 hours, and then it decreased but remained about twice the level after 48 hours (Fig 1C). The pattern of induction of the EF-1α gene was similar to that of the cyclin G gene (Fig 1D). In 2-2 cells, which express another mutant of p53 (not a temperature-sensitive protein),28 no significant changes were observed in the expression of the EF-1α gene (Fig 1C). The level of EF-1α of 1-2-3 cells cultured at 32°C for 6 hours was about five times of that at 37°C, and it continued to increase for 48 hours (Figs 1E and 1F ).
Identification of the EF-1α gene as an upregulated gene by wild-type p53 in the erythroleukemic cell line 1-2-3, which expresses only the temperature-sensitive p53 (Val-135).28 We identified bands with higher intensity at 32°C than that at 37°C by the DD method using a Delta RNA Fingerprinting Kit (Clontech). One band (D9) was confirmed by Northern blotting analysis (A). (B) Sequence comparison of the D9 clone with the mouse EF-1α gene (GenBank/EMBL m17878). (C) Time course of the expression of the EF-1α gene in 1-2-3 cells and 2-2 cells after the temperature-shift. (D) Comparison of the patterns of expression of several genes that have been suggested to be upregulated by wild-type p53. (E) Western blotting analysis of EF-1α protein. (F ) The pattern of induction of EF-1α protein in 1-2-3 cells after the temperature-shift.
Identification of the EF-1α gene as an upregulated gene by wild-type p53 in the erythroleukemic cell line 1-2-3, which expresses only the temperature-sensitive p53 (Val-135).28 We identified bands with higher intensity at 32°C than that at 37°C by the DD method using a Delta RNA Fingerprinting Kit (Clontech). One band (D9) was confirmed by Northern blotting analysis (A). (B) Sequence comparison of the D9 clone with the mouse EF-1α gene (GenBank/EMBL m17878). (C) Time course of the expression of the EF-1α gene in 1-2-3 cells and 2-2 cells after the temperature-shift. (D) Comparison of the patterns of expression of several genes that have been suggested to be upregulated by wild-type p53. (E) Western blotting analysis of EF-1α protein. (F ) The pattern of induction of EF-1α protein in 1-2-3 cells after the temperature-shift.
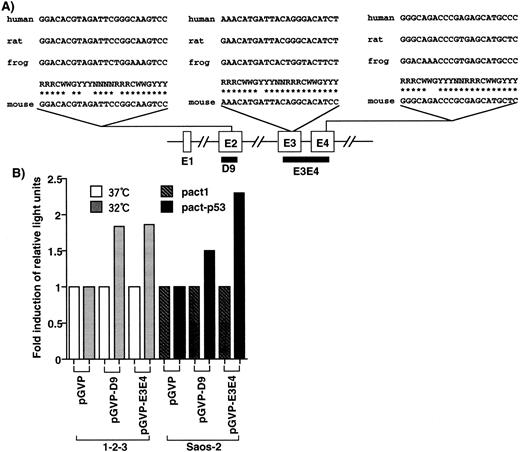
In the EF-1α gene, three candidates of p53-responsive elements, RRRC(A/T)(A/T)GYYY,36 were found in exon 2, exon 3, and exon 4 (Fig 2A), but no candidates were found in other genomic regions of the gene. D9, cloned by the DD method, corresponded to exon 2 and contained one such element. D9 and E3E4, corresponding to a genomic fragment of the mouse EF-1α gene with two p53-responsive elements, were cloned into the luciferase reporter plasmid pGVP. When cells were transfected with pGVP-D9, the luciferase activity of the cells with wild-type p53 (1-2-3 cells cultured at 32°C and Saos-2 cells transfected with pact-p53) was 1.5-fold to twofold of that of cells without wild-type p53 (1-2-3 cells 37°C and Saos-2 cells transfected with pact1; Fig 2B). In the case of pGVP-E3E4, the luciferase activity of the cells with wild-type p53 was twofold to 2.5-fold of that of cells without wild-type p53 (Fig 2B). Because E3E4 contains two p53-responsive elements, the inducibility might be slightly stronger than D9. In any case, these elements conferred p53-inducibility, although a few mismatches with the consensus were found. Moreover, the induction of luciferase activity was consistent with the level of induction of mRNA, as analyzed by Northern blotting. Therefore, upregulation of the EF-1α gene by p53 might be mediated by these elements.
Identification of p53-responsive elements in the EF-1α gene. (A) Elements homologous to the p53-responsive consensus (RRRCWWGYYY)36 were identified in exon 2 (4-base mismatch), exon 3 (1-base mismatch), and exon 4 (2-base mismatch), respectively. These elements were conserved in the human, rat, and frog EF-1α gene. D9 contained the consensus sequence in exon 2. (B) D9 and genomic region (E3E4) that contained the consensus elements conferred the p53-inducibility.
Identification of p53-responsive elements in the EF-1α gene. (A) Elements homologous to the p53-responsive consensus (RRRCWWGYYY)36 were identified in exon 2 (4-base mismatch), exon 3 (1-base mismatch), and exon 4 (2-base mismatch), respectively. These elements were conserved in the human, rat, and frog EF-1α gene. D9 contained the consensus sequence in exon 2. (B) D9 and genomic region (E3E4) that contained the consensus elements conferred the p53-inducibility.
The EF-1α gene encodes a polypeptide elongation factor that is an important component of the eukaryotic translation apparatus.30 Diphtheria toxin, which inactivates elongation factor-2 (EF-2), another component of the eukaryotic translation apparatus, mediates apoptosis of a human ovarian tumor cell line.37 Inhibition and/or abnormal regulation of polypeptide synthesis might be associated with apoptotic cell death.
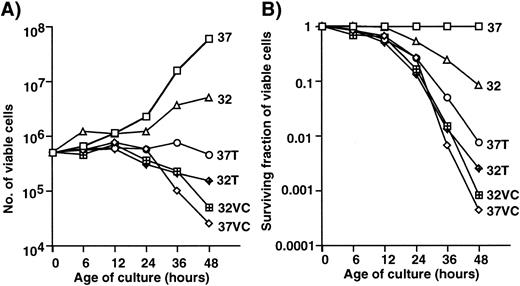
EF-1α has also been identified as a microtubule-severing factor.31 Because the rearrangement of microtubules occurs between interphase and mitosis, an unregulated elevated level of expression of EF-1α might be associated with cell death. The effects of microtubule-active drugs on cell growth and cell death were examined. Figure 3 shows the growth curves (A) and survival curves (B) of 1-2-3 cells with and without treatment with vincristine or Taxol at 32°C and at 37°C. Not much difference in terms of growth and survival was observed when vicristine-treated cells were cultured at 32°C and 37°C. Treatment with Taxol at 32°C was slightly more effective in inducing growth arrest and death than that at 37°C. Taxol stabilizes microtubules, vincristine inhibits the polymerization of microtubule, and EF-1α is associated with the microtubule severing. The functional difference between Taxol and EF-1α might result in the synergistic effect of Taxol at 32°C. The strong effect of vincristine might mask that of EF-1α because of the functional similarity between vincristine and EF-1α. Therefore, the effect of temperature-shift was minimal in cells treated with vincristine.
Effects of Taxol and vincristine (VC) on the growth and survival of 1-2-3 cells. Number (A) and surviving fraction (B) of viable cells with or without treatment with drugs. 37, Cells cultured at 37°C; 32, cells cultured at 32°C; 37T, cells treated with Taxol at 37°C; 32T, cells treated with Taxol at 32°C; 37VC, cells treated with vincristine at 37°C; 32VC, cells treated with vincristine at 32°C.
Effects of Taxol and vincristine (VC) on the growth and survival of 1-2-3 cells. Number (A) and surviving fraction (B) of viable cells with or without treatment with drugs. 37, Cells cultured at 37°C; 32, cells cultured at 32°C; 37T, cells treated with Taxol at 37°C; 32T, cells treated with Taxol at 32°C; 37VC, cells treated with vincristine at 37°C; 32VC, cells treated with vincristine at 32°C.
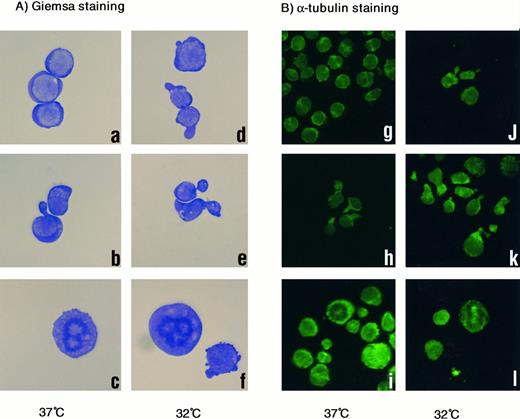
Morphologic analysis of treated cells at 32°C or 37°C also suggested a functional similarity between vincristine and EF-1α (Fig 4). Cells without treatment were round and appeared normal (Fig 4A, a), whereas cells cultured at 32°C and/or treated with vincristine had a distorted, necked shape (Fig 4A, b, d, and e). Neck-like regions were strongly stained with antibody against α-tubulin (Fig 4B, h, j, and k). Maldistribution of microtubules caused by severing associated with the unscheduled overexpression of EF-1α might result in distortion of the cytoskeleton. Therefore, cells became unable to maintain a normal cell shape and might have died. In Taxol treatment, metaphase-arrested cells with abnormal stabilization of microtubule were frequently observed (Fig 4A, c and f; and B, i and l).
(A) Giemsa staining of 1-2-3 cells. (a and d) No treatment at 37°C and 32°C; (b and e) treated with vincristine at 37°C and 32°C; and (c and f ) treated with Taxol at 37°C and 32°C. Cells were photographed at 200× magnification. (B) α-Tubulin staining. (g and j) No treatment at 37°C and 32°C; (h and k) treated with vincristine at 37°C and 32°C; (i and l) treated with Taxol at 37°C and 32°C. Cells were photographed at 100× magnification.
(A) Giemsa staining of 1-2-3 cells. (a and d) No treatment at 37°C and 32°C; (b and e) treated with vincristine at 37°C and 32°C; and (c and f ) treated with Taxol at 37°C and 32°C. Cells were photographed at 200× magnification. (B) α-Tubulin staining. (g and j) No treatment at 37°C and 32°C; (h and k) treated with vincristine at 37°C and 32°C; (i and l) treated with Taxol at 37°C and 32°C. Cells were photographed at 100× magnification.
Recently, the negative regulation of the microtubule-associated protein (MAP4) by p53 in p53-induced apoptosis was reported.38 Microtubule-associated protein must be one of targets for induction of apoptosis. Upregulation of the EF-1α gene by p53 may be enough to induce apoptosis without significant changes in the expression of the apoptosis-associated genes in 1-2-3 cells.28
The accumulated evidence that p53 also plays a role in G2/M checkpoint regulation39 40 suggests that EF-1α might be a key factor in the regulation of cell cycle at G2/M. The present finding that p53 upregulates the EF-1α gene provides new insights into the mechanism of the cell cycle and cell death, and it might also be relevant to attempts at cancer therapy.
ACKNOWLEDGMENT
The authors are very grateful to K. Sudo and S. Kobayashi for their assistance.
Supported in part by a grant-in-aid for the New Ten-Year Strategy for the Conquest of Cancer from the Ministry of Health and Welfare of Japan and by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science and Culture, Japan.
Address reprint requests to Mitsuo V. Kato, PhD, Laboratory of Molecular Oncology, Tsukuba Life Science Center, The Institute of Physical and Chemical Research (RIKEN), 3-1-1, Koyadai, Tsukuba, Ibaraki 305, Japan.